bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1974_orf1
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
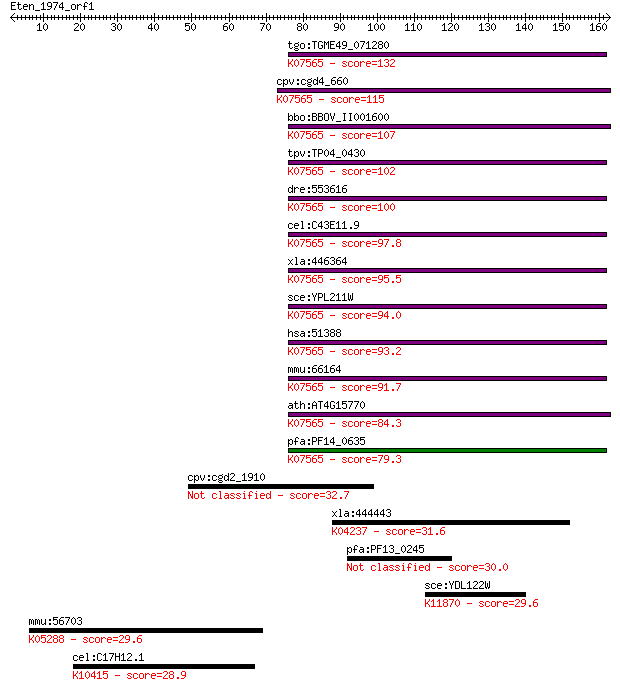
tgo:TGME49_071280 60S ribosome subunit biogenesis protein NIP7... 132 5e-31
cpv:cgd4_660 Nip7 like PUA domain containing protein involved ... 115 9e-26
bbo:BBOV_II001600 18.m06124; 60S ribosome subunit biogenesis p... 107 1e-23
tpv:TP04_0430 hypothetical protein; K07565 60S ribosome subuni... 102 7e-22
dre:553616 nip7, MGC110384, zgc:110384; nuclear import 7 homol... 100 3e-21
cel:C43E11.9 hypothetical protein; K07565 60S ribosome subunit... 97.8 1e-20
xla:446364 nip7, MGC83122; nuclear import 7 homolog; K07565 60... 95.5 6e-20
sce:YPL211W NIP7; Nip7p; K07565 60S ribosome subunit biogenesi... 94.0 2e-19
hsa:51388 NIP7, FLJ10296, HSPC031, KD93; nuclear import 7 homo... 93.2 3e-19
mmu:66164 Nip7, 1110017C15Rik, 6330509M23Rik, AA408773, AA4100... 91.7 9e-19
ath:AT4G15770 RNA binding / protein binding; K07565 60S riboso... 84.3 1e-16
pfa:PF14_0635 RNA binding protein, putative; K07565 60S riboso... 79.3 5e-15
cpv:cgd2_1910 phosphatidylinositol-4-phosphate 5-kinase, MORN ... 32.7 0.48
xla:444443 pard3, MGC83635, par-3, par3; par-3 partitioning de... 31.6 1.3
pfa:PF13_0245 conserved Plasmodium protein, unknown function 30.0 3.8
sce:YDL122W UBP1; Ubiquitin-specific protease that removes ubi... 29.6 4.2
mmu:56703 Pigo, FLJ00350, mFLJ00350; phosphatidylinositol glyc... 29.6 5.0
cel:C17H12.1 dyci-1; DYnein Chain, light Intermediate family m... 28.9 7.0
> tgo:TGME49_071280 60S ribosome subunit biogenesis protein NIP7,
putative ; K07565 60S ribosome subunit biogenesis protein
NIP7
Length=181
Score = 132 bits (332), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 59/86 (68%), Positives = 72/86 (83%), Gaps = 0/86 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MR LTE+E+KLV EKLSKF+G NL+ +++N DPHVFRLH DRVF++SEK+LK AGC+PR
Sbjct 1 MRQLTEEEVKLVFEKLSKFVGTNLMQIVDNQEDPHVFRLHHDRVFYMSEKVLKMAGCIPR 60
Query 136 KNLLSVGQCIGKITKGRKFRLGITAL 161
K+LL G CIGK TK RKFRL +TAL
Sbjct 61 KSLLLAGVCIGKFTKSRKFRLQVTAL 86
> cpv:cgd4_660 Nip7 like PUA domain containing protein involved
in ribosomal biogenesis ; K07565 60S ribosome subunit biogenesis
protein NIP7
Length=183
Score = 115 bits (287), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 52/90 (57%), Positives = 71/90 (78%), Gaps = 0/90 (0%)
Query 73 RLKMRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGC 132
++KMRPLTE+E K++ EKLS +LG+NLL L+E + +VFRLH+ VF++SE L KAA
Sbjct 1 KVKMRPLTEEETKILFEKLSNYLGNNLLSLLEREDESYVFRLHKGNVFYMSETLAKAASI 60
Query 133 VPRKNLLSVGQCIGKITKGRKFRLGITALH 162
VP+K+LLS+G C+GK +K KFRLGIT+L
Sbjct 61 VPKKSLLSMGTCLGKFSKNNKFRLGITSLQ 90
> bbo:BBOV_II001600 18.m06124; 60S ribosome subunit biogenesis
protein NIP7; K07565 60S ribosome subunit biogenesis protein
NIP7
Length=180
Score = 107 bits (268), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 46/89 (51%), Positives = 69/89 (77%), Gaps = 2/89 (2%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIEN--SSDPHVFRLHRDRVFFLSEKLLKAAGCV 133
MRPL+EDE +LV +KL+ ++G+ ++ ++ + S P VFRLHR+RV+++SE +LK +GC+
Sbjct 1 MRPLSEDEAQLVFQKLAAYIGNKVVDMVNDGDSGSPFVFRLHRERVYYMSESILKYSGCI 60
Query 134 PRKNLLSVGQCIGKITKGRKFRLGITALH 162
KNL+S G C+GK TK ++FRL ITALH
Sbjct 61 SHKNLISAGVCLGKFTKSKQFRLHITALH 89
> tpv:TP04_0430 hypothetical protein; K07565 60S ribosome subunit
biogenesis protein NIP7
Length=178
Score = 102 bits (253), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 44/87 (50%), Positives = 66/87 (75%), Gaps = 1/87 (1%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSS-DPHVFRLHRDRVFFLSEKLLKAAGCVP 134
MRPL+EDE+++VL+KL+ ++GD +L ++EN+ H FRLH +RV+++SE +LK G +
Sbjct 1 MRPLSEDEVQIVLKKLASYIGDKVLQMVENNQFGQHYFRLHNERVYYISENILKYVGSIS 60
Query 135 RKNLLSVGQCIGKITKGRKFRLGITAL 161
K L+S G C+GK TK ++FRL ITAL
Sbjct 61 SKKLISAGVCLGKFTKSKQFRLHITAL 87
> dre:553616 nip7, MGC110384, zgc:110384; nuclear import 7 homolog
(S. cerevisiae); K07565 60S ribosome subunit biogenesis
protein NIP7
Length=180
Score = 100 bits (248), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 48/86 (55%), Positives = 63/86 (73%), Gaps = 0/86 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MRPLT++E K + EKLSK++G+N+ LI+ + FRLH DRV+++SEK+LK A V R
Sbjct 1 MRPLTDEETKTMFEKLSKYIGENIKLLIDRPDGTYCFRLHNDRVYYMSEKILKLATNVSR 60
Query 136 KNLLSVGQCIGKITKGRKFRLGITAL 161
L+SVG C GK TK +KFRL ITAL
Sbjct 61 DKLVSVGTCFGKFTKTQKFRLHITAL 86
> cel:C43E11.9 hypothetical protein; K07565 60S ribosome subunit
biogenesis protein NIP7
Length=180
Score = 97.8 bits (242), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 44/86 (51%), Positives = 61/86 (70%), Gaps = 0/86 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MRPLTE+E LV KL+ F+GDN+ LI+ + + FR H++RV++ SE L++ A C+ R
Sbjct 1 MRPLTEEETSLVFAKLASFIGDNVSMLIDRNDGDYCFRNHKERVYYCSENLMRQAACIAR 60
Query 136 KNLLSVGQCIGKITKGRKFRLGITAL 161
+ LLS G C+GK TK +KF L ITAL
Sbjct 61 EPLLSFGTCLGKFTKSKKFHLQITAL 86
> xla:446364 nip7, MGC83122; nuclear import 7 homolog; K07565
60S ribosome subunit biogenesis protein NIP7
Length=180
Score = 95.5 bits (236), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 43/86 (50%), Positives = 62/86 (72%), Gaps = 0/86 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MRPLT++E + + EKLSK++G+N+ L++ + FRLH DRV+++SEK+LK A + R
Sbjct 1 MRPLTDEETRAMFEKLSKYIGENIKLLVDRPDGTYCFRLHNDRVYYVSEKILKLATNIAR 60
Query 136 KNLLSVGQCIGKITKGRKFRLGITAL 161
L+S G C GK TK +KFRL +TAL
Sbjct 61 DKLVSFGTCFGKFTKTQKFRLHVTAL 86
> sce:YPL211W NIP7; Nip7p; K07565 60S ribosome subunit biogenesis
protein NIP7
Length=181
Score = 94.0 bits (232), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 44/86 (51%), Positives = 62/86 (72%), Gaps = 0/86 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MR LTE+E K+V EKL+ ++G N+ L++N PHVFRL +DRV+++ + + K A V R
Sbjct 1 MRQLTEEETKVVFEKLAGYIGRNISFLVDNKELPHVFRLQKDRVYYVPDHVAKLATSVAR 60
Query 136 KNLLSVGQCIGKITKGRKFRLGITAL 161
NL+S+G C+GK TK KFRL IT+L
Sbjct 61 PNLMSLGICLGKFTKTGKFRLHITSL 86
> hsa:51388 NIP7, FLJ10296, HSPC031, KD93; nuclear import 7 homolog
(S. cerevisiae); K07565 60S ribosome subunit biogenesis
protein NIP7
Length=133
Score = 93.2 bits (230), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 41/86 (47%), Positives = 62/86 (72%), Gaps = 0/86 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MRPLTE+E +++ EK++K++G+NL L++ + FRLH DRV+++SEK++K A +
Sbjct 1 MRPLTEEETRVMFEKIAKYIGENLQLLVDRPDGTYCFRLHNDRVYYVSEKIMKLAANISG 60
Query 136 KNLLSVGQCIGKITKGRKFRLGITAL 161
L+S+G C GK TK KFRL +TAL
Sbjct 61 DKLVSLGTCFGKFTKTHKFRLHVTAL 86
> mmu:66164 Nip7, 1110017C15Rik, 6330509M23Rik, AA408773, AA410017,
PEachy; nuclear import 7 homolog (S. cerevisiae); K07565
60S ribosome subunit biogenesis protein NIP7
Length=133
Score = 91.7 bits (226), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 41/86 (47%), Positives = 61/86 (70%), Gaps = 0/86 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MRPLTE+E +++ EK++K++G+NL L++ + FRLH DRV+++SE +LK A +
Sbjct 1 MRPLTEEETRVMFEKIAKYIGENLQLLVDRPDGTYCFRLHNDRVYYVSEMMLKLAANISG 60
Query 136 KNLLSVGQCIGKITKGRKFRLGITAL 161
L+S+G C GK TK KFRL +TAL
Sbjct 61 DKLVSLGTCFGKFTKTHKFRLHVTAL 86
> ath:AT4G15770 RNA binding / protein binding; K07565 60S ribosome
subunit biogenesis protein NIP7
Length=187
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 44/95 (46%), Positives = 60/95 (63%), Gaps = 8/95 (8%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSD--------PHVFRLHRDRVFFLSEKLL 127
MRPL E E +V EKL KF+G+NL ++EN SD + FRL ++RV+++SE L+
Sbjct 1 MRPLDETETTVVFEKLFKFVGNNLKKIVENPSDEGPESTPGSYCFRLQKNRVYYVSEALV 60
Query 128 KAAGCVPRKNLLSVGQCIGKITKGRKFRLGITALH 162
K A + RKNL+S G CIGK T F L I +L+
Sbjct 61 KRATNISRKNLVSFGTCIGKYTHAGSFHLTIMSLN 95
> pfa:PF14_0635 RNA binding protein, putative; K07565 60S ribosome
subunit biogenesis protein NIP7
Length=179
Score = 79.3 bits (194), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 36/86 (41%), Positives = 58/86 (67%), Gaps = 0/86 (0%)
Query 76 MRPLTEDEIKLVLEKLSKFLGDNLLHLIENSSDPHVFRLHRDRVFFLSEKLLKAAGCVPR 135
MRPL + E +V +KLSKF+G+NLL ++ S++ ++ RLHR V+F+ + K A + +
Sbjct 1 MRPLNDQETMIVFKKLSKFVGNNLLTMLSYSNEEYILRLHRSNVYFVRADVAKQAESLNK 60
Query 136 KNLLSVGQCIGKITKGRKFRLGITAL 161
+L+S+G C+GK TK F + ITA+
Sbjct 61 NSLISMGICLGKFTKTNNFFIKITAI 86
> cpv:cgd2_1910 phosphatidylinositol-4-phosphate 5-kinase, MORN
beta hairpin repeats glycine-rich protein
Length=534
Score = 32.7 bits (73), Expect = 0.48, Method: Composition-based stats.
Identities = 20/50 (40%), Positives = 27/50 (54%), Gaps = 5/50 (10%)
Query 49 ACTSTSPLHWENRIIAQKPLAWGWRLKMRPLTEDEIKLVLEKLSKFLGDN 98
C S + H ENR I Q P R++ RPL E + K+ E ++LGDN
Sbjct 3 GCKSKADKHIENREIPQAP---ELRVEKRPLVELQDKITYE--GEWLGDN 47
> xla:444443 pard3, MGC83635, par-3, par3; par-3 partitioning
defective 3 homolog; K04237 partitioning defective protein 3
Length=1073
Score = 31.6 bits (70), Expect = 1.3, Method: Composition-based stats.
Identities = 22/71 (30%), Positives = 32/71 (45%), Gaps = 7/71 (9%)
Query 88 LEKLSKFLGDNLLHLIENSSDPHVFRLH-------RDRVFFLSEKLLKAAGCVPRKNLLS 140
LE+ S F D+++ L+E S+D +H R L K L+ G R+NL
Sbjct 215 LERTSSFSLDDMVKLVEVSNDGGPLGIHVVPYSARGGRTLGLLVKRLEKGGKAERENLFH 274
Query 141 VGQCIGKITKG 151
CI +I G
Sbjct 275 ENDCIVRINNG 285
> pfa:PF13_0245 conserved Plasmodium protein, unknown function
Length=595
Score = 30.0 bits (66), Expect = 3.8, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 20/28 (71%), Gaps = 2/28 (7%)
Query 92 SKFLGDNLLHLIENSSDPHVFRLHRDRV 119
+KF G LLHLI+ S D H++ L++ ++
Sbjct 319 NKFYG--LLHLIQKSRDKHIYSLYKKKI 344
> sce:YDL122W UBP1; Ubiquitin-specific protease that removes ubiquitin
from ubiquitinated proteins; cleaves at the C terminus
of ubiquitin fusions irrespective of their size; capable
of cleaving polyubiquitin chains (EC:3.1.2.15); K11870 ubiquitin
carboxyl-terminal hydrolase 1 [EC:3.1.2.15]
Length=809
Score = 29.6 bits (65), Expect = 4.2, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 113 RLHRDRVFFLSEKLLKAAGCVPRKNLL 139
+ +RD+ +F + LLKA PRKN+L
Sbjct 213 KYYRDKPYFKTNSLLKAMSKSPRKNIL 239
> mmu:56703 Pigo, FLJ00350, mFLJ00350; phosphatidylinositol glycan
anchor biosynthesis, class O; K05288 phosphatidylinositol
glycan, class O
Length=1101
Score = 29.6 bits (65), Expect = 5.0, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 36/65 (55%), Gaps = 16/65 (24%)
Query 6 LLLMLICSKTPLSKLLKSFSLTYI--EAQSNTLLFGSVEWYLLYSACTSTSPLHWENRII 63
L+L+LI +L FS +++ EA++ L GS+ ++L+ + LHWE +++
Sbjct 559 LILLLI-------RLATFFSDSFVVAEARATPFLLGSLVFFLV-------AQLHWEGQLL 604
Query 64 AQKPL 68
KPL
Sbjct 605 PPKPL 609
> cel:C17H12.1 dyci-1; DYnein Chain, light Intermediate family
member (dyci-1); K10415 dynein intermediate chain, cytosolic
Length=643
Score = 28.9 bits (63), Expect = 7.0, Method: Composition-based stats.
Identities = 12/49 (24%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 18 SKLLKSFSLTYIEAQSNTLLFGSVEWYLLYSACTSTSPLHWENRIIAQK 66
SK + + + Y +++ ++ F +LL + C S W+NR+ ++K
Sbjct 323 SKRMTAEFIVYCQSEIQSVAFARFHAHLLLAGCESGQICVWDNRLTSRK 371
Lambda K H
0.324 0.139 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3767900632
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40