bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2005_orf1
Length=187
Score E
Sequences producing significant alignments: (Bits) Value
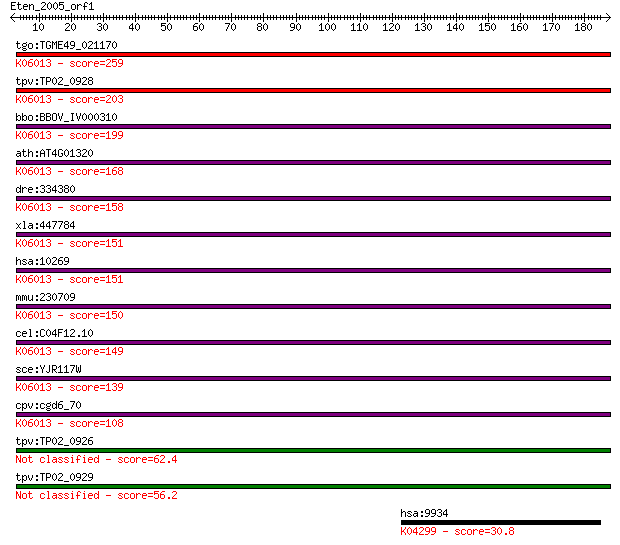
tgo:TGME49_021170 peptidase family M48 domain-containing prote... 259 5e-69
tpv:TP02_0928 CAAX prenyl protease 1; K06013 STE24 endopeptida... 203 3e-52
bbo:BBOV_IV000310 21.m02775; CAAX metallo endopeptidase; K0601... 199 3e-51
ath:AT4G01320 ATSTE24; endopeptidase/ metalloendopeptidase; K0... 168 1e-41
dre:334380 zmpste24, fi45a02, wu:fi45a02, zgc:55655; zinc meta... 158 8e-39
xla:447784 zmpste24, MGC85351; zinc metallopeptidase (STE24 ho... 151 1e-36
hsa:10269 ZMPSTE24, FACE-1, FACE1, FLJ14968, HGPS, PRO1, STE24... 151 1e-36
mmu:230709 Zmpste24, A530043O15Rik, D030046F19, FACE1, Face-1,... 150 2e-36
cel:C04F12.10 fce-1; Farnesylated-proteins Converting Enzyme (... 149 4e-36
sce:YJR117W STE24, AFC1, PIO2; Highly conserved zinc metallopr... 139 6e-33
cpv:cgd6_70 hypothetical protein ; K06013 STE24 endopeptidase ... 108 1e-23
tpv:TP02_0926 hypothetical protein 62.4 9e-10
tpv:TP02_0929 hypothetical protein 56.2 6e-08
hsa:9934 P2RY14, GPR105, KIAA0001, P2Y14; purinergic receptor ... 30.8 2.5
> tgo:TGME49_021170 peptidase family M48 domain-containing protein
(EC:3.4.24.84); K06013 STE24 endopeptidase [EC:3.4.24.84]
Length=432
Score = 259 bits (661), Expect = 5e-69, Method: Compositional matrix adjust.
Identities = 120/185 (64%), Positives = 146/185 (78%), Gaps = 0/185 (0%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FNKKT +F+KDKLL LGLT IG P+ CA WLIKWGG++F+ WLWGFSV TT ++ +
Sbjct 146 FNKKTLGIFVKDKLLSLGLTGLIGGPLACAAIWLIKWGGKSFYLWLWGFSVATTIALMFV 205
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
YPNFIAPLFNKF+ L D ELR KI LA++L+FPL ++YEMD SKRS HSNAYFYGFW
Sbjct 206 YPNFIAPLFNKFEPLKDEELRGKICELAKKLDFPLTQLYEMDNSKRSGHSNAYFYGFWWS 265
Query 123 KRIVIFDTLLHLPHDQILAILGHELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMNNDHL 182
KRIV++DTLLHLPHDQILAILGHE+GHW+ +H + + F+ LF TFYL+G VM +D L
Sbjct 266 KRIVLYDTLLHLPHDQILAILGHEMGHWKRSHTTKMMAATFLQLFCTFYLFGLVMTSDAL 325
Query 183 YKCFG 187
+ FG
Sbjct 326 FDSFG 330
> tpv:TP02_0928 CAAX prenyl protease 1; K06013 STE24 endopeptidase
[EC:3.4.24.84]
Length=444
Score = 203 bits (516), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 96/185 (51%), Positives = 134/185 (72%), Gaps = 0/185 (0%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FNKKT LF+KD LL L L IG P+LCA+ +L+ WGGE F+F+++GF V+ F M+++
Sbjct 160 FNKKTYKLFVKDLLLTLLLQCVIGGPVLCALIFLVNWGGELFYFYVFGFIVVFNFIMLIV 219
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
YP IAPLFNKF+ L D ELR+ I+ LA +++FPL E+ +MDGSKRSSHSNAY YG W++
Sbjct 220 YPELIAPLFNKFEPLHDEELRNDIENLARKVDFPLKEIKQMDGSKRSSHSNAYLYGLWKF 279
Query 123 KRIVIFDTLLHLPHDQILAILGHELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMNNDHL 182
K++VI+DTLL +I++++ HELGHW+ H + L F NLF F+L+ K +N ++
Sbjct 280 KKVVIYDTLLKQDRKEIVSVVSHELGHWKHKHVPKMLTFSFANLFAMFFLFKKFKDNKNM 339
Query 183 YKCFG 187
Y FG
Sbjct 340 YNSFG 344
> bbo:BBOV_IV000310 21.m02775; CAAX metallo endopeptidase; K06013
STE24 endopeptidase [EC:3.4.24.84]
Length=448
Score = 199 bits (507), Expect = 3e-51, Method: Compositional matrix adjust.
Identities = 97/185 (52%), Positives = 129/185 (69%), Gaps = 0/185 (0%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FNKKT LF KD L+ GL IGAP+L V +L+ WGGE F+ ++ F + M+VI
Sbjct 164 FNKKTIRLFFKDLLISFGLQIVIGAPVLSIVIFLVNWGGEYFYLYVGVFVAVFYLFMMVI 223
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
YP+FIAPLFNKF+ L D EL+ I+ LA++L FPL E+ MDGSKRS+HSN YFYGFW +
Sbjct 224 YPDFIAPLFNKFEPLNDNELKKDIEDLAQKLKFPLREIKLMDGSKRSNHSNMYFYGFWWF 283
Query 123 KRIVIFDTLLHLPHDQILAILGHELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMNNDHL 182
K+IV++DTLL P QI+AI HE+GHW+ NH ++ LL + LF FYL+G N+ +
Sbjct 284 KKIVMYDTLLKQPKQQIIAITAHEMGHWKCNHTLKLLLFSYTQLFAMFYLFGLFKNDAGM 343
Query 183 YKCFG 187
++ FG
Sbjct 344 FESFG 348
> ath:AT4G01320 ATSTE24; endopeptidase/ metalloendopeptidase;
K06013 STE24 endopeptidase [EC:3.4.24.84]
Length=424
Score = 168 bits (425), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 83/187 (44%), Positives = 122/187 (65%), Gaps = 2/187 (1%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FNK+T +FI+D + G L+ +G PI+ A+ ++++ GG +LW F + + M+ I
Sbjct 140 FNKQTIWMFIRDMIKGTFLSVILGPPIVAAIIFIVQKGGPYLAIYLWAFMFILSLVMMTI 199
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
YP IAPLFNKF L D +LR KI+ LA L FPL +++ +DGS RSSHSNAY YGF++
Sbjct 200 YPVLIAPLFNKFTPLPDGDLREKIEKLASSLKFPLKKLFVVDGSTRSSHSNAYMYGFFKN 259
Query 123 KRIVIFDTLLHL--PHDQILAILGHELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMNND 180
KRIV++DTL+ D+I+A++ HELGHW++NH + V + F+ F Y V N+
Sbjct 260 KRIVLYDTLIQQCKNEDEIVAVIAHELGHWKLNHTTYSFIAVQILAFLQFGGYTLVRNST 319
Query 181 HLYKCFG 187
L++ FG
Sbjct 320 DLFRSFG 326
> dre:334380 zmpste24, fi45a02, wu:fi45a02, zgc:55655; zinc metallopeptidase,
STE24 homolog (EC:3.4.24.84); K06013 STE24 endopeptidase
[EC:3.4.24.84]
Length=468
Score = 158 bits (400), Expect = 8e-39, Method: Compositional matrix adjust.
Identities = 81/221 (36%), Positives = 130/221 (58%), Gaps = 36/221 (16%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FN++T F+KD L +T I P+ + ++IK GG+ F + W F+ + + ++ I
Sbjct 150 FNQQTLGFFLKDALKKFAVTQCILVPVTSLLLYIIKIGGDYFFIYAWLFTFIVSLILVTI 209
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
Y ++IAPLF+KF L D ELRS+I+++A+ ++FPL ++Y ++GSKRSSHSNAYFYGF++
Sbjct 210 YADYIAPLFDKFTPLPDGELRSEIESMAKSISFPLTKLYVVEGSKRSSHSNAYFYGFFKN 269
Query 123 KRIVIFDTLLH------------------------------------LPHDQILAILGHE 146
KRIV+FDTLL + ++LA+LGHE
Sbjct 270 KRIVLFDTLLEDYSPLNQSGEKEPGTGEENEAVVNESKAKPKNKKQGCSNPEVLAVLGHE 329
Query 147 LGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMNNDHLYKCFG 187
LGHW++ H ++ +++ MN F+ F+L+ ++ L+ FG
Sbjct 330 LGHWKLGHTVKNIVISQMNSFLCFFLFAVLIGRKELFMAFG 370
> xla:447784 zmpste24, MGC85351; zinc metallopeptidase (STE24
homolog); K06013 STE24 endopeptidase [EC:3.4.24.84]
Length=465
Score = 151 bits (382), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 76/218 (34%), Positives = 129/218 (59%), Gaps = 33/218 (15%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FN++T F KD + + +T I P+ + ++IK GG+ F + W F+++ + ++ I
Sbjct 150 FNQQTLGFFFKDAVKKILVTQCILLPVASLLLYIIKMGGDYFFIYAWLFTLVVSLVLVTI 209
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
Y ++IAPLF+KF L + +L+ I+ +A+ ++FPL +VY ++GSKRSSHSNAYFYGF++
Sbjct 210 YADYIAPLFDKFTPLSEGDLKEAIENMAKSIDFPLTKVYVVEGSKRSSHSNAYFYGFFKN 269
Query 123 KRIVIFDTLLH---------------------------------LPHDQILAILGHELGH 149
KRIV+FDTLL + ++LA+LGHELGH
Sbjct 270 KRIVLFDTLLEDYSPLNKEGTDDTSGNENTELKSKVKNLNKKQGCNNQEVLAVLGHELGH 329
Query 150 WQMNHFMQRLLLVFMNLFVTFYLYGKVMNNDHLYKCFG 187
W++ H ++ +++ +N F+ F+L+ ++ L++ FG
Sbjct 330 WKLGHTVKNIVISQVNSFLCFFLFAVLIGRKELFEAFG 367
> hsa:10269 ZMPSTE24, FACE-1, FACE1, FLJ14968, HGPS, PRO1, STE24,
Ste24p; zinc metallopeptidase (STE24 homolog, S. cerevisiae)
(EC:3.4.24.84); K06013 STE24 endopeptidase [EC:3.4.24.84]
Length=475
Score = 151 bits (382), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 77/223 (34%), Positives = 130/223 (58%), Gaps = 38/223 (17%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FN++T F+KD + +T I P+ + ++IK GG+ F + W F+++ + ++ I
Sbjct 155 FNQQTLGFFMKDAIKKFVVTQCILLPVSSLLLYIIKIGGDYFFIYAWLFTLVVSLVLVTI 214
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
Y ++IAPLF+KF L + +L+ +I+ +A+ ++FPL +VY ++GSKRSSHSNAYFYGF++
Sbjct 215 YADYIAPLFDKFTPLPEGKLKEEIEVMAKSIDFPLTKVYVVEGSKRSSHSNAYFYGFFKN 274
Query 123 KRIVIFDTLLH--------------------------------------LPHDQILAILG 144
KRIV+FDTLL ++++LA+LG
Sbjct 275 KRIVLFDTLLEEYSVLNKDIQEDSGMEPRNEEEGNSEEIKAKVKNKKQGCKNEEVLAVLG 334
Query 145 HELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMNNDHLYKCFG 187
HELGHW++ H ++ +++ MN F+ F+L+ ++ L+ FG
Sbjct 335 HELGHWKLGHTVKNIIISQMNSFLCFFLFAVLIGRKELFAAFG 377
> mmu:230709 Zmpste24, A530043O15Rik, D030046F19, FACE1, Face-1,
MADB, STE24, Ste24p; zinc metallopeptidase, STE24 homolog
(S. cerevisiae) (EC:3.4.24.84); K06013 STE24 endopeptidase
[EC:3.4.24.84]
Length=475
Score = 150 bits (379), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 78/223 (34%), Positives = 131/223 (58%), Gaps = 38/223 (17%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FN +T F+KD + +T I P+ + ++IK GG+ F + W F+++ + ++ I
Sbjct 155 FNHQTLEFFMKDAIKKFIVTQCILLPVSALLLYIIKIGGDYFFIYAWLFTLVVSLVLVTI 214
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
Y ++IAPLF+KF L + +L+ +I+ +A+ ++FPL +VY ++GSKRSSHSNAYFYGF++
Sbjct 215 YADYIAPLFDKFTPLPEGKLKQEIEVMAKSIDFPLTKVYVVEGSKRSSHSNAYFYGFFKN 274
Query 123 KRIVIFDTLLH---LP-----------------------------------HDQILAILG 144
KRIV+FDTLL +P ++++LA+LG
Sbjct 275 KRIVLFDTLLEEYSVPNKDNQEESGMEARNEGEGDSEEVKAKVKNKKQGCKNEEVLAVLG 334
Query 145 HELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMNNDHLYKCFG 187
HELGHW++ H ++ +++ MN F+ F+L+ ++ L+ FG
Sbjct 335 HELGHWKLGHTVKNIIISQMNSFLCFFLFAVLIGRRELFAAFG 377
> cel:C04F12.10 fce-1; Farnesylated-proteins Converting Enzyme
(FACE) family member (fce-1); K06013 STE24 endopeptidase [EC:3.4.24.84]
Length=442
Score = 149 bits (377), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 81/213 (38%), Positives = 126/213 (59%), Gaps = 28/213 (13%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FNK+T + DK+ + + A+ PI+ + W+I GG F ++W F + ++ I
Sbjct 131 FNKQTIGFYFVDKIKKMLVGFALTMPIVYGIEWIIVNGGPYFFVYIWLFVSVVVLLLMTI 190
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
YP FIAPLF+K+ L D +L++KI+ LA L++PL E+Y ++GSKRS+HSNAY YGFW+
Sbjct 191 YPTFIAPLFDKYFPLPDGDLKTKIEQLAASLSYPLTELYVVNGSKRSAHSNAYMYGFWKN 250
Query 123 KRIVIFDTLLH----------------------------LPHDQILAILGHELGHWQMNH 154
KRIV++DTLL + +D+++A+LGHELGHW + H
Sbjct 251 KRIVLYDTLLSGAEKEKVHELYVAAGEKIEETENDKKRGMNNDEVVAVLGHELGHWALWH 310
Query 155 FMQRLLLVFMNLFVTFYLYGKVMNNDHLYKCFG 187
+ L++ +NLF +F ++G + LY+ FG
Sbjct 311 TLINLVITEVNLFFSFAVFGYFYKWEALYQGFG 343
> sce:YJR117W STE24, AFC1, PIO2; Highly conserved zinc metalloprotease
that functions in two steps of A-factor maturation,
C-terminal CAAX proteolysis and the first step of N-terminal
proteolytic processing; contains multiple transmembrane spans
(EC:3.4.24.84); K06013 STE24 endopeptidase [EC:3.4.24.84]
Length=453
Score = 139 bits (350), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 77/187 (41%), Positives = 112/187 (59%), Gaps = 2/187 (1%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FNK T L+I D + L L AIG PIL + +F +++ F + + I
Sbjct 153 FNKLTVQLWITDMIKSLTLAYAIGGPILYLFLKIFDKFPTDFLWYIMVFLFVVQILAMTI 212
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGF-WR 121
P FI P+FNKF L D EL+ I++LA+++ FPL +++ +DGSKRSSHSNAYF G +
Sbjct 213 IPVFIMPMFNKFTPLEDGELKKSIESLADRVGFPLDKIFVIDGSKRSSHSNAYFTGLPFT 272
Query 122 WKRIVIFDTLLHL-PHDQILAILGHELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMNND 180
KRIV+FDTL++ D+I A+L HE+GHWQ NH + ++ ++ F+ F L+ + N
Sbjct 273 SKRIVLFDTLVNSNSTDEITAVLAHEIGHWQKNHIVNMVIFSQLHTFLIFSLFTSIYRNT 332
Query 181 HLYKCFG 187
Y FG
Sbjct 333 SFYNTFG 339
> cpv:cgd6_70 hypothetical protein ; K06013 STE24 endopeptidase
[EC:3.4.24.84]
Length=432
Score = 108 bits (270), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 65/185 (35%), Positives = 105/185 (56%), Gaps = 0/185 (0%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
FN T +FI D++ L G +L + ++ G+ F+ ++ F +I
Sbjct 144 FNNSTLKIFIMDQIKSGLLVTVFGTILLSVMIYIANNTGKYFYVYIALVQFGFIFIFSII 203
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
YP I P+FNK + ++EL KI L + +NFPL +Y+MD S RS+H NA+F G ++
Sbjct 204 YPIIIVPIFNKLTPVENQELAEKISKLCKDVNFPLKNLYQMDASLRSNHGNAFFSGAFKS 263
Query 123 KRIVIFDTLLHLPHDQILAILGHELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMNNDHL 182
K I+++DT+L P D+I+AI+GHE+GHW+ + L F+ FVT +++ + + L
Sbjct 264 KSIILYDTILDFPQDEIVAIIGHEIGHWKNWDNYKLLFFSFIQTFVTLFIFHLTFSWNGL 323
Query 183 YKCFG 187
Y FG
Sbjct 324 YLSFG 328
> tpv:TP02_0926 hypothetical protein
Length=449
Score = 62.4 bits (150), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 43/189 (22%), Positives = 82/189 (43%), Gaps = 9/189 (4%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
F KT +F K L + + WL K NF + GF+ L F +++
Sbjct 162 FKTKTRFVFFKQYFLCYAFYVLALTGLASGLTWLQKLSKSNFR--VNGFAFLVVFKTVLV 219
Query 63 YPNFIAPLF----NKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYG 118
F PL +K L D ELR ++D + ++L V+ + + + + +G
Sbjct 220 ---FAVPLLLRLKHKLSPLADPELRREVDTMGKKLGLSSKNVHVVSHNTAHTDGVSLSWG 276
Query 119 FWRWKRIVIFDTLLHLPHDQILAILGHELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMN 178
F ++K + ++ + L ++A++ H GH++ +HF++ L + +LY
Sbjct 277 FCKFKHAYLNESYVTLGKPSVMALVAHTFGHFKHHHFLKSFLFDLTKVTFFLFLYDHFKG 336
Query 179 NDHLYKCFG 187
+ L+K F
Sbjct 337 DSALFKSFS 345
> tpv:TP02_0929 hypothetical protein
Length=458
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 40/185 (21%), Positives = 81/185 (43%), Gaps = 1/185 (0%)
Query 3 FNKKTAILFIKDKLLGLGLTAAIGAPILCAVNWLIKWGGENFHFWLWGFSVLTTFGMIVI 62
F ++ +F + L + + + W+ K+ NF + F V+ M ++
Sbjct 162 FKTRSRCVFFRQYFLTYVFYLLVFTGLASGLTWVQKFSKSNFRVNGFAFLVVFKTAMALV 221
Query 63 YPNFIAPLFNKFKVLGDRELRSKIDALAEQLNFPLCEVYEMDGSKRSSHSNAYFYGFWRW 122
P F++ L +K L D ELR ++D + ++L V+ + S +H +GF +
Sbjct 222 VPLFLS-LEHKLSPLPDPELRKEVDTMGKKLGLSSKNVHVVSSSTAPAHGLTLSWGFCMF 280
Query 123 KRIVIFDTLLHLPHDQILAILGHELGHWQMNHFMQRLLLVFMNLFVTFYLYGKVMNNDHL 182
K + ++ + L LA++ GH++ +HF++ LL +L+ + L
Sbjct 281 KHAYLNESYVTLGKPSALALVAVNFGHFKHHHFLKTFLLDLAKDTFFLFLFDHFKGDAAL 340
Query 183 YKCFG 187
+K F
Sbjct 341 FKSFN 345
> hsa:9934 P2RY14, GPR105, KIAA0001, P2Y14; purinergic receptor
P2Y, G-protein coupled, 14; K04299 purinergic receptor P2Y,
G protein-coupled, 14
Length=338
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 38/65 (58%), Gaps = 6/65 (9%)
Query 123 KRIVIFDTLLHLPHDQILAILGHE-LGHWQMNHFMQRL--LLVFMNLFVTFYLYGKVMNN 179
K IVI D ++ L ILG LG WQ+N F+ R+ +L ++N++V+ +G +++
Sbjct 61 KNIVIADFVMSLTFP--FKILGDSGLGPWQLNVFVCRVSAVLFYVNMYVSIVFFG-LISF 117
Query 180 DHLYK 184
D YK
Sbjct 118 DRYYK 122
Lambda K H
0.331 0.145 0.490
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5235419772
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40