bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2017_orf2
Length=256
Score E
Sequences producing significant alignments: (Bits) Value
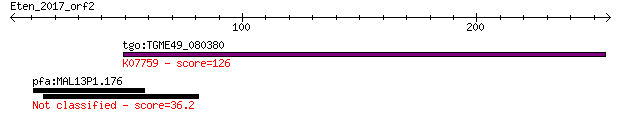
tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143); K0... 126 6e-29
pfa:MAL13P1.176 PfRh2b; reticulocyte binding protein 2, homolog B 36.2 0.11
> tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=553
Score = 126 bits (317), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 75/211 (35%), Positives = 113/211 (53%), Gaps = 17/211 (8%)
Query 49 AILLPQNPQGWQQVQQILAEVEAGKVPKTAGEYKQLFTSILTAAGNWRVDKPLRI---GP 105
AIL + W + + IL V +G P + L +L +GN R +K RI G
Sbjct 47 AILRTPVEEDWGKTKYILNFVASGFGPADPQQLASLQKRLLKLSGNIRTEKNKRILQRGL 106
Query 106 LTHHLATHKNLSKSLLPFLASLARRLPDFFPSGIRHLGRDNPAVHLRRIEVLALLAALFF 165
L + LPF+A+L R+ + FPSG++++ +NP VHLR+I+V L+AA F
Sbjct 107 LNYLEENPGKFFSHDLPFMATLVMRIDELFPSGLQYITPENPQVHLRKIQVFTLIAAAFL 166
Query 166 GIVYPRQRLLLQQQLQQQQQQQQPLQMNAPDMHYRGFFERPPKYQSLLQYFQQMQQTVGS 225
G++ QR LL Q++ + N DM YRGF ER PK+QS+L YF M++ +G+
Sbjct 167 GVIPHNQRALL-----AHHQKKFVMNQNKLDMFYRGFMERKPKFQSVLIYFASMRERLGN 221
Query 226 CWQQMLQRG--DVGPLLQRGRVCCGWCSSSS 254
CW +M+++G D+ P C G C S+
Sbjct 222 CWNEMVKKGFVDMAP-------CTGTCICST 245
> pfa:MAL13P1.176 PfRh2b; reticulocyte binding protein 2, homolog
B
Length=3179
Score = 36.2 bits (82), Expect = 0.11, Method: Composition-based stats.
Identities = 11/47 (23%), Positives = 34/47 (72%), Gaps = 0/47 (0%)
Query 11 KISKRRRKRLESRKQEQQRQQQQQQQQQSEREQQQGLRAILLPQNPQ 57
++ ++ ++RLE KQEQ +++++ ++++ E++QQ+ ++ + + P+
Sbjct 2676 ELKRQEQERLEREKQEQLQKEEELRKKEQEKQQQRNIQELEEQKKPE 2722
Score = 30.0 bits (66), Expect = 7.9, Method: Composition-based stats.
Identities = 24/99 (24%), Positives = 43/99 (43%), Gaps = 33/99 (33%)
Query 15 RRRKRLESRKQEQ-----------------------QRQQQQ----------QQQQQSER 41
+ ++RLE KQEQ +RQ+Q+ Q+Q++ ER
Sbjct 2628 QEKERLEREKQEQLKKEALKKQEQERQEQQQKEEALKRQEQERLQKEEELKRQEQERLER 2687
Query 42 EQQQGLRAILLPQNPQGWQQVQQILAEVEAGKVPKTAGE 80
E+Q+ L+ + + +Q Q+ + E+E K P+ E
Sbjct 2688 EKQEQLQKEEELRKKEQEKQQQRNIQELEEQKKPEIINE 2726
Lambda K H
0.324 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9340616888
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40