bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2042_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
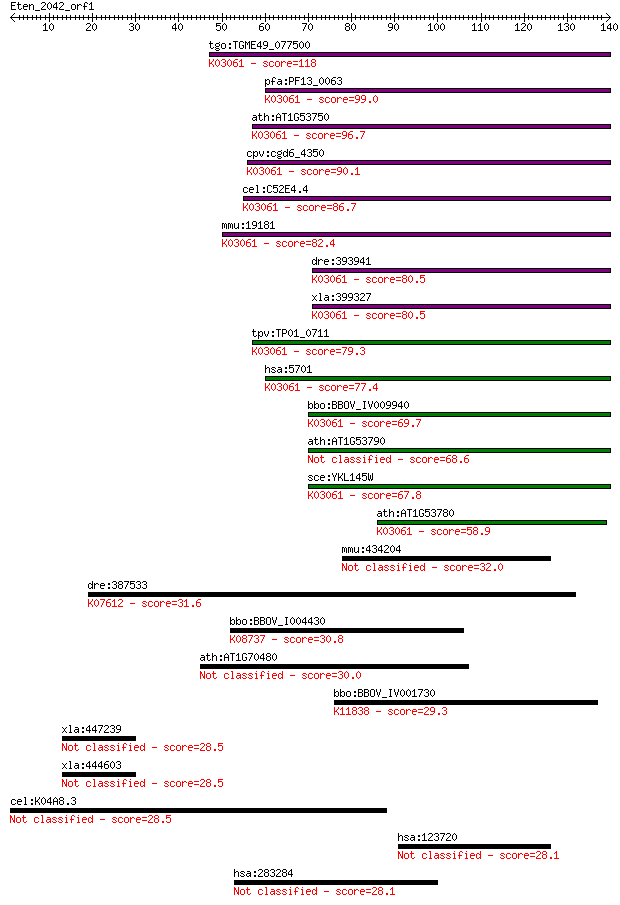
tgo:TGME49_077500 26S proteasome AAA-ATPase subunit RPT1, puta... 118 5e-27
pfa:PF13_0063 26S proteasome regulatory subunit 7, putative; K... 99.0 4e-21
ath:AT1G53750 RPT1A; RPT1A (REGULATORY PARTICLE TRIPLE-A 1A); ... 96.7 2e-20
cpv:cgd6_4350 26S proteasome regulatory subunit 7 (RPT1)-like.... 90.1 2e-18
cel:C52E4.4 rpt-1; proteasome Regulatory Particle, ATPase-like... 86.7 2e-17
mmu:19181 Psmc2; proteasome (prosome, macropain) 26S subunit, ... 82.4 4e-16
dre:393941 psmc2, MGC63995, zgc:63995; proteasome (prosome, ma... 80.5 1e-15
xla:399327 psmc2, xMSS1; proteasome (prosome, macropain) 26S s... 80.5 1e-15
tpv:TP01_0711 26S proteasome regulatory subunit 7; K03061 26S ... 79.3 3e-15
hsa:5701 PSMC2, MGC3004, MSS1, Nbla10058, S7; proteasome (pros... 77.4 1e-14
bbo:BBOV_IV009940 23.m06337; 26S protease regulatory subunit 7... 69.7 3e-12
ath:AT1G53790 F-box family protein 68.6 6e-12
sce:YKL145W RPT1, CIM5, YTA3; One of six ATPases of the 19S re... 67.8 1e-11
ath:AT1G53780 ATP binding / ATPase/ hydrolase/ nucleoside-trip... 58.9 4e-09
mmu:434204 Whamm, BB081391, KIAA1971, MGC51670, MGC58752, Whdc... 32.0 0.62
dre:387533 gja3, Cx44, cx46, cx48.5; gap junction protein, alp... 31.6 0.83
bbo:BBOV_I004430 19.m02191; DNA repair protein; K08737 DNA mis... 30.8 1.4
ath:AT1G70480 hypothetical protein 30.0 2.2
bbo:BBOV_IV001730 21.m03027; ubiquitin carboxyl-terminal hydro... 29.3 3.7
xla:447239 ttyh1, MGC84458, ttyh1-A; tweety homolog 1 28.5
xla:444603 MGC84077, ttyh1-B; protein tweety homolog 1-B 28.5 6.9
cel:K04A8.3 hypothetical protein 28.5 7.1
hsa:123720 WHAMM, KIAA1971, WHDC1; WAS protein homolog associa... 28.1 9.1
hsa:283284 IGSF22, FLJ37794; immunoglobulin superfamily, membe... 28.1 9.5
> tgo:TGME49_077500 26S proteasome AAA-ATPase subunit RPT1, putative
; K03061 26S proteasome regulatory subunit T1
Length=476
Score = 118 bits (296), Expect = 5e-27, Method: Composition-based stats.
Identities = 59/95 (62%), Positives = 71/95 (74%), Gaps = 2/95 (2%)
Query 47 ANPSSSRPKG--DESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQN 104
A ++ RPK D + +D+ PLD ADI+ILKSYGLSPYA + K+L+ DIK +
Sbjct 12 APEAAERPKAKKDAAHEDEEPKKTIPLDDADINILKSYGLSPYAFAIKRLDTDIKSLTEK 71
Query 105 ISKLCGVRESDTGLSLPSQWDLAADKRLMQEQPLQ 139
I+KLCGVRESDTGL PSQWDLAADK+LMQEQPLQ
Sbjct 72 ITKLCGVRESDTGLCQPSQWDLAADKQLMQEQPLQ 106
> pfa:PF13_0063 26S proteasome regulatory subunit 7, putative;
K03061 26S proteasome regulatory subunit T1
Length=420
Score = 99.0 bits (245), Expect = 4e-21, Method: Composition-based stats.
Identities = 49/81 (60%), Positives = 62/81 (76%), Gaps = 2/81 (2%)
Query 60 EKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDTGLS 119
E++ NT + P LD DI+ILKSYG PY+ + KK+E DI L NI+KLCGVRESDTGL
Sbjct 2 EEETNTQSKP-LDDEDINILKSYGSGPYSKTIKKVEGDISGLLVNINKLCGVRESDTGLC 60
Query 120 LPSQWDLAADKRLM-QEQPLQ 139
LP+QWDL DK+++ +EQPLQ
Sbjct 61 LPNQWDLQLDKQMLNEEQPLQ 81
> ath:AT1G53750 RPT1A; RPT1A (REGULATORY PARTICLE TRIPLE-A 1A);
ATPase; K03061 26S proteasome regulatory subunit T1
Length=426
Score = 96.7 bits (239), Expect = 2e-20, Method: Composition-based stats.
Identities = 47/84 (55%), Positives = 61/84 (72%), Gaps = 4/84 (4%)
Query 57 DESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDT 116
DE +KN P PLD DI +LK+YGL PY+ KK+E +IK + I+ LCG++ESDT
Sbjct 7 DEIRDEKN---PRPLDEDDIALLKTYGLGPYSAPIKKVEKEIKDLAKKINDLCGIKESDT 63
Query 117 GLSLPSQWDLAADKRLMQ-EQPLQ 139
GL+ PSQWDL +DK++MQ EQPLQ
Sbjct 64 GLAPPSQWDLVSDKQMMQEEQPLQ 87
> cpv:cgd6_4350 26S proteasome regulatory subunit 7 (RPT1)-like.
AAA atpase ; K03061 26S proteasome regulatory subunit T1
Length=432
Score = 90.1 bits (222), Expect = 2e-18, Method: Composition-based stats.
Identities = 44/85 (51%), Positives = 60/85 (70%), Gaps = 5/85 (5%)
Query 56 GDESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESD 115
G + E+D++ PLD DIDI++ YG+ PY K++E ++K + ISKL +RESD
Sbjct 13 GGKEEEDQSR----PLDQVDIDIMRKYGMGPYVSKIKQVETEVKDLMSAISKLNSIRESD 68
Query 116 TGLSLPSQWDLAADKRLMQ-EQPLQ 139
TGL+ PSQWDLAAD++ MQ EQPLQ
Sbjct 69 TGLAPPSQWDLAADRQNMQMEQPLQ 93
> cel:C52E4.4 rpt-1; proteasome Regulatory Particle, ATPase-like
family member (rpt-1); K03061 26S proteasome regulatory subunit
T1
Length=435
Score = 86.7 bits (213), Expect = 2e-17, Method: Composition-based stats.
Identities = 46/86 (53%), Positives = 60/86 (69%), Gaps = 4/86 (4%)
Query 55 KGDESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRES 114
K D E++KN A LD DI +LK YG PYA K L+ADI+ L+ +++L GV+ES
Sbjct 14 KDDTKEEEKNFQA---LDEGDIAVLKRYGQGPYAEQLKTLDADIENCLKKVNELSGVKES 70
Query 115 DTGLSLPSQWDLAADKRLM-QEQPLQ 139
DTGL+ P+ WD+AADK+ M QEQPLQ
Sbjct 71 DTGLAPPALWDIAADKQAMQQEQPLQ 96
> mmu:19181 Psmc2; proteasome (prosome, macropain) 26S subunit,
ATPase 2; K03061 26S proteasome regulatory subunit T1
Length=475
Score = 82.4 bits (202), Expect = 4e-16, Method: Composition-based stats.
Identities = 44/91 (48%), Positives = 62/91 (68%), Gaps = 4/91 (4%)
Query 50 SSSRPKGDESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLC 109
+ R +E + DK A LD DI +LK+YG S Y+ K++E DI+Q L+ I++L
Sbjct 49 ADQRKTKEEEKDDKPIRA---LDEGDIALLKTYGQSTYSRQIKQVEDDIQQLLKKINELT 105
Query 110 GVRESDTGLSLPSQWDLAADKRLMQ-EQPLQ 139
G++ESDTGL+ P+ WDLAADK+ +Q EQPLQ
Sbjct 106 GIKESDTGLAPPALWDLAADKQTLQSEQPLQ 136
> dre:393941 psmc2, MGC63995, zgc:63995; proteasome (prosome,
macropain) 26S subunit, ATPase 2; K03061 26S proteasome regulatory
subunit T1
Length=433
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 39/70 (55%), Positives = 54/70 (77%), Gaps = 1/70 (1%)
Query 71 LDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWDLAADK 130
LD DI +LK+YG S Y+ K++E DI+Q L+ I++L G++ESDTGL+ P+ WDLAADK
Sbjct 25 LDEGDIALLKTYGQSTYSRQIKQVEDDIQQLLKKINELTGIKESDTGLAPPALWDLAADK 84
Query 131 RLMQ-EQPLQ 139
+ +Q EQPLQ
Sbjct 85 QTLQSEQPLQ 94
> xla:399327 psmc2, xMSS1; proteasome (prosome, macropain) 26S
subunit, ATPase, 2; K03061 26S proteasome regulatory subunit
T1
Length=433
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 39/70 (55%), Positives = 54/70 (77%), Gaps = 1/70 (1%)
Query 71 LDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWDLAADK 130
LD DI +LK+YG S Y+ K++E DI+Q L+ I++L G++ESDTGL+ P+ WDLAADK
Sbjct 25 LDEGDIALLKTYGQSTYSRQIKQVEDDIQQLLKKINELTGIKESDTGLAPPALWDLAADK 84
Query 131 RLMQ-EQPLQ 139
+ +Q EQPLQ
Sbjct 85 QTLQSEQPLQ 94
> tpv:TP01_0711 26S proteasome regulatory subunit 7; K03061 26S
proteasome regulatory subunit T1
Length=425
Score = 79.3 bits (194), Expect = 3e-15, Method: Composition-based stats.
Identities = 40/84 (47%), Positives = 55/84 (65%), Gaps = 4/84 (4%)
Query 57 DESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDT 116
++SE+ K T PL ++ ILKSYG+ PYA + + DIK+ + I+KL GV+ESDT
Sbjct 6 EDSEEKKETV---PLSDTEVKILKSYGVGPYAAPIRAADNDIKEVINRINKLSGVKESDT 62
Query 117 GLSLPSQWDLAADKRLMQE-QPLQ 139
GL+ P WDL D+R +QE PLQ
Sbjct 63 GLNPPHMWDLVLDQRSLQEGAPLQ 86
> hsa:5701 PSMC2, MGC3004, MSS1, Nbla10058, S7; proteasome (prosome,
macropain) 26S subunit, ATPase, 2; K03061 26S proteasome
regulatory subunit T1
Length=141
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 41/81 (50%), Positives = 58/81 (71%), Gaps = 1/81 (1%)
Query 60 EKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDTGLS 119
E +K+ LD DI +LK+YG S Y+ K++E DI+Q L+ I++L G++ESDTGL+
Sbjct 14 EDEKDDKPIRALDEGDIALLKTYGQSTYSRQIKQVEDDIQQLLKKINELTGIKESDTGLA 73
Query 120 LPSQWDLAADKRLMQ-EQPLQ 139
P+ WDLAADK+ +Q EQPLQ
Sbjct 74 PPALWDLAADKQTLQSEQPLQ 94
> bbo:BBOV_IV009940 23.m06337; 26S protease regulatory subunit
7; K03061 26S proteasome regulatory subunit T1
Length=425
Score = 69.7 bits (169), Expect = 3e-12, Method: Composition-based stats.
Identities = 35/71 (49%), Positives = 48/71 (67%), Gaps = 1/71 (1%)
Query 70 PLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWDLAAD 129
PL+ A+I ILKS+G+ PY S + E IK +Q I+KL G++ESDTGL+ WDL D
Sbjct 16 PLNDAEISILKSFGIGPYTESISESEKGIKTVVQRINKLSGIKESDTGLNPLHMWDLMYD 75
Query 130 KRLMQEQ-PLQ 139
++ +QE PLQ
Sbjct 76 QQALQEGVPLQ 86
> ath:AT1G53790 F-box family protein
Length=461
Score = 68.6 bits (166), Expect = 6e-12, Method: Composition-based stats.
Identities = 35/71 (49%), Positives = 49/71 (69%), Gaps = 8/71 (11%)
Query 70 PLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWDLAAD 129
PLD DI ++K+YGL PY+ KK+E +IK+ + I+ LCG++ESDTGL+ S
Sbjct 9 PLDEDDIALVKTYGLVPYSAPIKKVEKEIKELTEKINDLCGIKESDTGLAPLS------- 61
Query 130 KRLMQE-QPLQ 139
K++MQE QPLQ
Sbjct 62 KQMMQEGQPLQ 72
> sce:YKL145W RPT1, CIM5, YTA3; One of six ATPases of the 19S
regulatory particle of the 26S proteasome involved in the degradation
of ubiquitinated substrates; required for optimal
CDC20 transcription; interacts with Rpn12p and the E3 ubiquitin-protein
ligase Ubr1p; K03061 26S proteasome regulatory subunit
T1
Length=467
Score = 67.8 bits (164), Expect = 1e-11, Method: Composition-based stats.
Identities = 34/71 (47%), Positives = 44/71 (61%), Gaps = 1/71 (1%)
Query 70 PLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWDLAAD 129
PL DI +LKSYG +PYA K+ E D+K I + GV+ESDTGL+ WD+ D
Sbjct 28 PLTEGDIQVLKSYGAAPYAAKLKQTENDLKDIEARIKEKAGVKESDTGLAPSHLWDIMGD 87
Query 130 K-RLMQEQPLQ 139
+ RL +E PLQ
Sbjct 88 RQRLGEEHPLQ 98
> ath:AT1G53780 ATP binding / ATPase/ hydrolase/ nucleoside-triphosphatase/
nucleotide binding / peptidyl-prolyl cis-trans
isomerase; K03061 26S proteasome regulatory subunit T1
Length=598
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 28/54 (51%), Positives = 40/54 (74%), Gaps = 2/54 (3%)
Query 86 PYAGSTKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWDLAADKRLMQ-EQPL 138
PY+ KK+E +I + + I L G++ESDTGL+ P+QWDL +DK++MQ EQPL
Sbjct 205 PYSARIKKVEKEINELAEKICNL-GIKESDTGLAPPNQWDLVSDKQMMQEEQPL 257
> mmu:434204 Whamm, BB081391, KIAA1971, MGC51670, MGC58752, Whdc1,
mKIAA1971; WAS protein homolog associated with actin, golgi
membranes and microtubules
Length=793
Score = 32.0 bits (71), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 78 ILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWD 125
+ G+ P ST LE I++ L+ I K+ E D P++WD
Sbjct 745 VHTGQGVDPGKKSTSDLERSIREALERIKKVSADSEEDNDEPSPTEWD 792
> dre:387533 gja3, Cx44, cx46, cx48.5; gap junction protein, alpha
3; K07612 gap junction protein, alpha 3
Length=434
Score = 31.6 bits (70), Expect = 0.83, Method: Composition-based stats.
Identities = 30/120 (25%), Positives = 54/120 (45%), Gaps = 7/120 (5%)
Query 19 SCVAALLRVCCPRWLHYLLSLLGAAREMANPSSSRPKGDESEKDKNTAAPP----PLDAA 74
+CV+ LL + L + G E A S P+ DE+E + APP P D
Sbjct 228 ACVSLLLNLLEIYHLGWKKVKQGMTNEFAPDRESLPEADEAEPESPRTAPPTLSYPPDYT 287
Query 75 DIDILKSYGLSPY-AGSTKKLEAD-IKQQLQNISKLCGVRESDTGLSLPSQW-DLAADKR 131
++ + L P A ST + + D ++++L+ S ++ L+ W +LA +++
Sbjct 288 EVAVAGGAFLQPVSAPSTAEFKMDPLREELEESSPFYISNNNNHRLAAEQNWANLATEQQ 347
> bbo:BBOV_I004430 19.m02191; DNA repair protein; K08737 DNA mismatch
repair protein MSH6
Length=1313
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 28/54 (51%), Gaps = 1/54 (1%)
Query 52 SRPKGDESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQNI 105
S PK D++E D T AP P+ + DI +S L GS KL K + ++I
Sbjct 126 SIPK-DDNENDATTVAPEPVASIQPDICESSALVNRMGSIAKLSHTKKSRAKDI 178
> ath:AT1G70480 hypothetical protein
Length=338
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 30/62 (48%), Gaps = 15/62 (24%)
Query 45 EMANPSSSRPKGDESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIKQQLQN 104
EM NP G ++E++K D A LS + G ++KL+ +K QL+N
Sbjct 18 EMGNP------GSDTERNKGNVVSSVFDPA---------LSRFVGFSQKLQTRLKSQLKN 62
Query 105 IS 106
++
Sbjct 63 LT 64
> bbo:BBOV_IV001730 21.m03027; ubiquitin carboxyl-terminal hydrolase;
K11838 ubiquitin carboxyl-terminal hydrolase 7 [EC:3.1.2.15]
Length=1446
Score = 29.3 bits (64), Expect = 3.7, Method: Composition-based stats.
Identities = 17/61 (27%), Positives = 29/61 (47%), Gaps = 6/61 (9%)
Query 76 IDILKSYGLSPYAGSTKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWDLAADKRLMQE 135
+ I+ YG P++ + L A L N+ +RE L S+W + ADK +++
Sbjct 922 VPIISDYGNGPFSYQSGDLGAGAPTSLPNLDHTSVMRE------LQSRWAMVADKEVIRY 975
Query 136 Q 136
Q
Sbjct 976 Q 976
> xla:447239 ttyh1, MGC84458, ttyh1-A; tweety homolog 1
Length=449
Score = 28.5 bits (62), Expect = 6.9, Method: Composition-based stats.
Identities = 10/17 (58%), Positives = 14/17 (82%), Gaps = 1/17 (5%)
Query 13 LCTYTWSCVAALLRVCC 29
+C TWSCVAA++ +CC
Sbjct 85 VCCVTWSCVAAVI-ICC 100
> xla:444603 MGC84077, ttyh1-B; protein tweety homolog 1-B
Length=451
Score = 28.5 bits (62), Expect = 6.9, Method: Composition-based stats.
Identities = 10/17 (58%), Positives = 14/17 (82%), Gaps = 1/17 (5%)
Query 13 LCTYTWSCVAALLRVCC 29
+C TWSCVAA++ +CC
Sbjct 85 VCCVTWSCVAAVI-ICC 100
> cel:K04A8.3 hypothetical protein
Length=117
Score = 28.5 bits (62), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 37/87 (42%), Gaps = 0/87 (0%)
Query 1 TLVKELLLLLGDLCTYTWSCVAALLRVCCPRWLHYLLSLLGAAREMANPSSSRPKGDESE 60
++ + L L + L + + + P+ H + EM+ S+ R + DE +
Sbjct 6 SINRGLGLGMAQLGAFARMGMMINVSAMGPQDGHANGGVGEGGIEMSRKSTWRSENDEIK 65
Query 61 KDKNTAAPPPLDAADIDILKSYGLSPY 87
K+TA P LD ID + G S Y
Sbjct 66 TRKSTAFEPDLDFDMIDFRQPSGSSQY 92
> hsa:123720 WHAMM, KIAA1971, WHDC1; WAS protein homolog associated
with actin, golgi membranes and microtubules
Length=809
Score = 28.1 bits (61), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 17/35 (48%), Gaps = 0/35 (0%)
Query 91 TKKLEADIKQQLQNISKLCGVRESDTGLSLPSQWD 125
T LE IK LQ I ++ E D+ P QWD
Sbjct 774 TSDLERSIKAALQRIKRVSADSEEDSDEQDPGQWD 808
> hsa:283284 IGSF22, FLJ37794; immunoglobulin superfamily, member
22
Length=1326
Score = 28.1 bits (61), Expect = 9.5, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 53 RPKGDESEKDKNTAAPPPLDAADIDILKSYGLSPYAGSTKKLEADIK 99
R KG ESE A PP +D + ++ L ++ ++ G T ++ +
Sbjct 586 RAKGTESEASVFIADPPTIDPSVLEALAAHAITVKVGHTAHIKVPFR 632
Lambda K H
0.316 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2487377096
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40