bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2047_orf2
Length=113
Score E
Sequences producing significant alignments: (Bits) Value
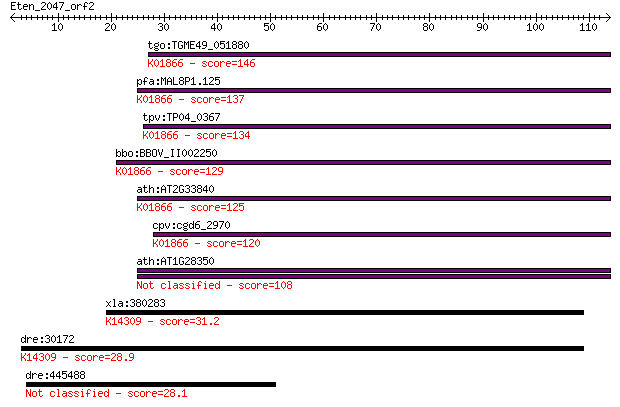
tgo:TGME49_051880 tyrosyl-tRNA synthetase, putative (EC:6.1.1.... 146 2e-35
pfa:MAL8P1.125 tyrosyl-tRNA synthetase, putative (EC:6.1.1.1);... 137 1e-32
tpv:TP04_0367 tyrosyl-tRNA synthetase (EC:6.1.1.1); K01866 tyr... 134 9e-32
bbo:BBOV_II002250 18.m06182; tyrosyl-tRNA synthetase; K01866 t... 129 2e-30
ath:AT2G33840 tRNA synthetase class I (W and Y) family protein... 125 3e-29
cpv:cgd6_2970 tyrosyl-tRNA synthetase (tyrosyl-tRNA ligase; Ty... 120 1e-27
ath:AT1G28350 ATP binding / aminoacyl-tRNA ligase/ nucleotide ... 108 4e-24
xla:380283 nup93, An4, MGC53870, dye, xnup93; nucleoporin 93kD... 31.2 0.95
dre:30172 nup93, dye; nucleoporin 93; K14309 nuclear pore comp... 28.9 3.7
dre:445488 morc2, zgc:92010; MORC family CW-type zinc finger 2 28.1
> tgo:TGME49_051880 tyrosyl-tRNA synthetase, putative (EC:6.1.1.1);
K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=427
Score = 146 bits (368), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 64/87 (73%), Positives = 77/87 (88%), Gaps = 0/87 (0%)
Query 27 KVGEYFVEVWKSAGMQMQNVQFLWASEEIGKDPETYWNLVMNIAKANSISRIKRCSQIMG 86
KVGEYF+EVWK+AGM MQNV+FLWASEEI ++ + YW VM IA++ +I+R+KRCSQIMG
Sbjct 169 KVGEYFIEVWKAAGMDMQNVRFLWASEEINRNSDQYWVQVMQIARSFTITRVKRCSQIMG 228
Query 87 RTEGDDQPAAQILYPCMQCADIFYLNA 113
R EGD+QPAAQI+YPCMQCADIFYL A
Sbjct 229 RQEGDEQPAAQIMYPCMQCADIFYLGA 255
> pfa:MAL8P1.125 tyrosyl-tRNA synthetase, putative (EC:6.1.1.1);
K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=373
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 57/89 (64%), Positives = 75/89 (84%), Gaps = 0/89 (0%)
Query 25 FRKVGEYFVEVWKSAGMQMQNVQFLWASEEIGKDPETYWNLVMNIAKANSISRIKRCSQI 84
+KVG YF+EVWKS GM M+NVQFLWASEEI K P YW+LV++I+++ +I+R+KRC +I
Sbjct 113 IKKVGSYFIEVWKSCGMNMENVQFLWASEEINKKPNEYWSLVLDISRSFNINRMKRCLKI 172
Query 85 MGRTEGDDQPAAQILYPCMQCADIFYLNA 113
MGR+EG++ +QILYPCMQCADIF+LN
Sbjct 173 MGRSEGEENYCSQILYPCMQCADIFFLNV 201
> tpv:TP04_0367 tyrosyl-tRNA synthetase (EC:6.1.1.1); K01866 tyrosyl-tRNA
synthetase [EC:6.1.1.1]
Length=427
Score = 134 bits (336), Expect = 9e-32, Method: Compositional matrix adjust.
Identities = 53/88 (60%), Positives = 75/88 (85%), Gaps = 0/88 (0%)
Query 26 RKVGEYFVEVWKSAGMQMQNVQFLWASEEIGKDPETYWNLVMNIAKANSISRIKRCSQIM 85
R VGEYF+ +WK+AGM +V+FLWAS+E+ K+P+ YW L M+I+++ +I+R+KRCSQ +
Sbjct 166 RVVGEYFIHIWKAAGMNTDSVKFLWASDEVDKNPDLYWRLSMDISRSFNITRMKRCSQAL 225
Query 86 GRTEGDDQPAAQILYPCMQCADIFYLNA 113
GRTEGDDQP+AQ+LYP +QCADIF++ A
Sbjct 226 GRTEGDDQPSAQLLYPAIQCADIFFIGA 253
> bbo:BBOV_II002250 18.m06182; tyrosyl-tRNA synthetase; K01866
tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=418
Score = 129 bits (325), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 53/93 (56%), Positives = 73/93 (78%), Gaps = 0/93 (0%)
Query 21 QMAFFRKVGEYFVEVWKSAGMQMQNVQFLWASEEIGKDPETYWNLVMNIAKANSISRIKR 80
M R VGEYF VW+++GM M V+FLWASEEI K+P+ YW LVM+I+++ +I+R+KR
Sbjct 143 NMENIRLVGEYFKHVWRASGMDMNAVKFLWASEEINKNPDLYWRLVMDISRSFNITRLKR 202
Query 81 CSQIMGRTEGDDQPAAQILYPCMQCADIFYLNA 113
CS+ +GR +GD++P A +LYP MQCADIFY+ A
Sbjct 203 CSEALGRADGDNRPGASLLYPAMQCADIFYIGA 235
> ath:AT2G33840 tRNA synthetase class I (W and Y) family protein;
K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=385
Score = 125 bits (314), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 56/89 (62%), Positives = 67/89 (75%), Gaps = 0/89 (0%)
Query 25 FRKVGEYFVEVWKSAGMQMQNVQFLWASEEIGKDPETYWNLVMNIAKANSISRIKRCSQI 84
R VGEYF E+WK+AGM V+FLW+SEEI + YW LVM+IA+ N + RI RC QI
Sbjct 125 IRVVGEYFQEIWKAAGMDNDKVEFLWSSEEINSKADKYWPLVMDIARKNKLPRILRCVQI 184
Query 85 MGRTEGDDQPAAQILYPCMQCADIFYLNA 113
MGR+E D+ AAQILYPCMQCADIF+L A
Sbjct 185 MGRSETDELSAAQILYPCMQCADIFFLEA 213
> cpv:cgd6_2970 tyrosyl-tRNA synthetase (tyrosyl-tRNA ligase;
TyrRS). class-I aaRS ; K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=376
Score = 120 bits (300), Expect = 1e-27, Method: Composition-based stats.
Identities = 53/87 (60%), Positives = 66/87 (75%), Gaps = 1/87 (1%)
Query 28 VGEYFVEVWKSAGMQMQNVQFLWASEEI-GKDPETYWNLVMNIAKANSISRIKRCSQIMG 86
VGEYFV +WK+AGM M NV+F+WAS+ I G+D YW V +I++ +I+RIKRC QIMG
Sbjct 118 VGEYFVHIWKAAGMDMTNVRFVWASDFINGEDSNEYWLRVFDISRKFNITRIKRCCQIMG 177
Query 87 RTEGDDQPAAQILYPCMQCADIFYLNA 113
R E D+QP A + YPCMQCADIF L A
Sbjct 178 RQENDEQPCASVFYPCMQCADIFQLKA 204
> ath:AT1G28350 ATP binding / aminoacyl-tRNA ligase/ nucleotide
binding / tyrosine-tRNA ligase
Length=776
Score = 108 bits (270), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 49/89 (55%), Positives = 66/89 (74%), Gaps = 0/89 (0%)
Query 25 FRKVGEYFVEVWKSAGMQMQNVQFLWASEEIGKDPETYWNLVMNIAKANSISRIKRCSQI 84
R VGEYF E++++AGM +NV+FLW+S+EI + YW LVM+IA NS+++IKRC I
Sbjct 139 IRVVGEYFKEIFQAAGMNSENVEFLWSSDEINAKGDEYWPLVMDIACRNSLAQIKRCMPI 198
Query 85 MGRTEGDDQPAAQILYPCMQCADIFYLNA 113
MG +E ++ AA ILY CMQCAD F+L A
Sbjct 199 MGLSENEELSAAHILYVCMQCADTFFLEA 227
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 53/89 (59%), Positives = 64/89 (71%), Gaps = 0/89 (0%)
Query 25 FRKVGEYFVEVWKSAGMQMQNVQFLWASEEIGKDPETYWNLVMNIAKANSISRIKRCSQI 84
R VGEYF E+W++ GM V+FLWAS+EI YW LVM+IA+ N++ RI RC QI
Sbjct 517 IRVVGEYFKEIWQAGGMNNDKVEFLWASDEINGKGSKYWPLVMDIARRNNLRRILRCGQI 576
Query 85 MGRTEGDDQPAAQILYPCMQCADIFYLNA 113
MGR+E + AAQILYPCMQCADIF L A
Sbjct 577 MGRSETEVLSAAQILYPCMQCADIFLLEA 605
> xla:380283 nup93, An4, MGC53870, dye, xnup93; nucleoporin 93kDa;
K14309 nuclear pore complex protein Nup93
Length=820
Score = 31.2 bits (69), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 45/94 (47%), Gaps = 15/94 (15%)
Query 19 QQQMAFFRKVGEYFVEVWKSAGM--QMQNVQFLWASEEIGKDPETYWNLVMNIAKANSIS 76
Q QMAF R+ Y + +K+ + N+Q ++G P TY NLV S
Sbjct 250 QMQMAFVRQALNYLEQSYKNYTLISVFANLQ----QAQLGGVPGTY-NLV------RSFL 298
Query 77 RIKRCSQIMGRTEGDDQ--PAAQILYPCMQCADI 108
I+ + + G +G+ + P ++Y CM+C D+
Sbjct 299 NIRLPTTVPGLQDGEIEGYPVWALIYYCMRCGDL 332
> dre:30172 nup93, dye; nucleoporin 93; K14309 nuclear pore complex
protein Nup93
Length=820
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 26/108 (24%), Positives = 49/108 (45%), Gaps = 11/108 (10%)
Query 3 LLLLESREQQQQEQQQQQQMAFFRKVGEYFVEVWKSAGMQMQNVQFLWASEEIGKDPETY 62
+LL+ +++ + QMAF R+ ++ +K+ + V ++G P TY
Sbjct 234 VLLVPAKDTLKSRVSVDMQMAFVRQALQFLENSYKN--YTLVTVFGNLHQAQLGGVPGTY 291
Query 63 WNLVMNIAKANSISRIKRCSQIMGRTEGD--DQPAAQILYPCMQCADI 108
LV S IK + + GR +G+ P ++Y C++C D+
Sbjct 292 -QLVC------SFLNIKLPTPLPGRQDGEVEGHPVWALIYFCLRCGDL 332
> dre:445488 morc2, zgc:92010; MORC family CW-type zinc finger
2
Length=1035
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 11/47 (23%), Positives = 21/47 (44%), Gaps = 0/47 (0%)
Query 4 LLLESREQQQQEQQQQQQMAFFRKVGEYFVEVWKSAGMQMQNVQFLW 50
L+LE +Q ++ R +G+Y + WK G+ + + W
Sbjct 419 LVLEPTHNKQDFADAKEYRHLLRAMGDYLAQYWKDIGIAQKGIVKFW 465
Lambda K H
0.321 0.131 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2052546600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40