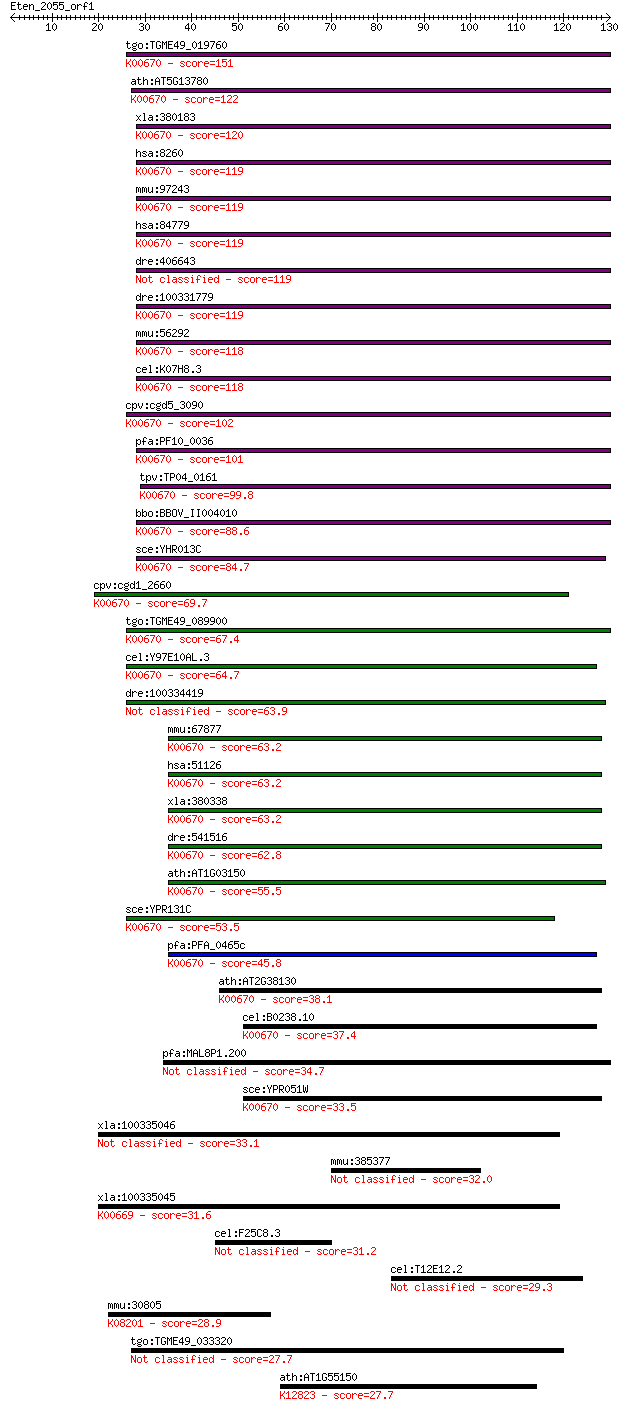
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2055_orf1
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_019760 N-terminal acetyltransferase complex subunit... 151 5e-37
ath:AT5G13780 GCN5-related N-acetyltransferase, putative; K006... 122 2e-28
xla:380183 naa10, MGC52756, ard1, ard1a, dxs707, te2; N(alpha)... 120 9e-28
hsa:8260 NAA10, ARD1, ARD1A, DXS707, MGC71248, TE2; N(alpha)-a... 119 2e-27
mmu:97243 Naa11, Ard1b, Ard2, C80008; N(alpha)-acetyltransfera... 119 2e-27
hsa:84779 NAA11, ARD1B, ARD2, MGC10646, hARD2; N(alpha)-acetyl... 119 2e-27
dre:406643 naa10, ard1a, hm:zeh0223, wu:fc66b08, zgc:63981; N(... 119 3e-27
dre:100331779 alpha-N-acetyltransferase 1A-like; K00670 peptid... 119 3e-27
mmu:56292 Naa10, 2310039H09Rik, Ard1, Ard1a, Te2; N(alpha)-ace... 118 4e-27
cel:K07H8.3 hypothetical protein; K00670 peptide alpha-N-acety... 118 5e-27
cpv:cgd5_3090 N-acetyltransferase subunit ARD1 ; K00670 peptid... 102 4e-22
pfa:PF10_0036 N-acetyltransferase, putative; K00670 peptide al... 101 6e-22
tpv:TP04_0161 N-acetyltransferase (EC:2.3.1.-); K00670 peptide... 99.8 2e-21
bbo:BBOV_II004010 18.m06331; N-acetyltransferase; K00670 pepti... 88.6 4e-18
sce:YHR013C ARD1, NAA10; Ard1p (EC:2.3.1.88); K00670 peptide a... 84.7 7e-17
cpv:cgd1_2660 n-terminal acetyltransferase complex ard1 ; K006... 69.7 2e-12
tgo:TGME49_089900 N-acetyltransferase 5, putative (EC:2.3.1.88... 67.4 1e-11
cel:Y97E10AL.3 hypothetical protein; K00670 peptide alpha-N-ac... 64.7 8e-11
dre:100334419 N-acetyltransferase 5-like 63.9 1e-10
mmu:67877 Naa20, 1500004D14Rik, 2900026I01Rik, AU041458, D2Ert... 63.2 2e-10
hsa:51126 NAA20, NAT3, NAT5, dJ1002M8.1; N(alpha)-acetyltransf... 63.2 2e-10
xla:380338 naa20, MGC53877, nat3, nat5; N(alpha)-acetyltransfe... 63.2 2e-10
dre:541516 naa20, im:7150897, nat5, zgc:110819; N(alpha)-acety... 62.8 2e-10
ath:AT1G03150 GCN5-related N-acetyltransferase (GNAT) family p... 55.5 4e-08
sce:YPR131C NAT3, NAA20; Catalytic subunit of the NatB N-termi... 53.5 1e-07
pfa:PFA_0465c N-terminal acetyltransferase, putative (EC:2.3.1... 45.8 3e-05
ath:AT2G38130 ATMAK3; ATMAK3; N-acetyltransferase; K00670 pept... 38.1 0.006
cel:B0238.10 hypothetical protein; K00670 peptide alpha-N-acet... 37.4 0.011
pfa:MAL8P1.200 Acetyltransferase, putative 34.7 0.067
sce:YPR051W MAK3, NAA30; Catalytic subunit of N-terminal acety... 33.5 0.19
xla:100335046 aanat, aanat-b, aanat1, nat4, snat; arylalkylami... 33.1 0.24
mmu:385377 Pnma5, KIAA1934, mKIAA1934; paraneoplastic antigen ... 32.0 0.53
xla:100335045 aanat-a; arylalkylamine N-acetyltransferase a (E... 31.6 0.60
cel:F25C8.3 unc-80; UNCoordinated family member (unc-80) 31.2 0.89
cel:T12E12.2 hypothetical protein 29.3 2.8
mmu:30805 Slc22a4, Octn1; solute carrier family 22 (organic ca... 28.9 4.2
tgo:TGME49_033320 hypothetical protein 27.7 8.6
ath:AT1G55150 DEAD box RNA helicase, putative (RH20); K12823 A... 27.7 10.0
> tgo:TGME49_019760 N-terminal acetyltransferase complex subunit
ARD1, putative (EC:2.3.1.88); K00670 peptide alpha-N-acetyltransferase
[EC:2.3.1.88]
Length=221
Score = 151 bits (381), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 69/104 (66%), Positives = 89/104 (85%), Gaps = 2/104 (1%)
Query 26 SLVRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKL 85
+ +R +DVFDLF MQ+ NF+NLPENY+MKY+FFH++SWPQL +V+ D QGKLVGYVLAKL
Sbjct 2 ATLRRADVFDLFAMQNANFINLPENYIMKYYFFHAVSWPQLLSVSHDSQGKLVGYVLAKL 61
Query 86 EDENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
E+++ HGHVTSVAV+RQ+RKLGLASKLM ++Q AM++VF
Sbjct 62 EEDDPTDH--HGHVTSVAVLRQSRKLGLASKLMNMTQHAMEEVF 103
> ath:AT5G13780 GCN5-related N-acetyltransferase, putative; K00670
peptide alpha-N-acetyltransferase [EC:2.3.1.88]
Length=192
Score = 122 bits (307), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 56/103 (54%), Positives = 77/103 (74%), Gaps = 3/103 (2%)
Query 27 LVRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLE 86
+R + V DL MQ N + LPENY MKY+ +H LSWPQL VA+D G++VGYVLAK+E
Sbjct 3 CIRRATVDDLLAMQACNLMCLPENYQMKYYLYHILSWPQLLYVAEDYNGRIVGYVLAKME 62
Query 87 DENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+E+ + HGH+TS+AV+R +RKLGLA+KLM +Q AM++V+
Sbjct 63 EESNEC---HGHITSLAVLRTHRKLGLATKLMTAAQAAMEQVY 102
> xla:380183 naa10, MGC52756, ard1, ard1a, dxs707, te2; N(alpha)-acetyltransferase
10, NatA catalytic subunit (EC:2.3.1.88);
K00670 peptide alpha-N-acetyltransferase [EC:2.3.1.88]
Length=237
Score = 120 bits (301), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 56/102 (54%), Positives = 73/102 (71%), Gaps = 2/102 (1%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+R + DL MQH N + LPENY MKY+F+H LSWPQL +A+D GK+VGYVLAK+E+
Sbjct 3 IRNARPEDLMNMQHCNLLCLPENYQMKYYFYHGLSWPQLSYIAEDENGKIVGYVLAKMEE 62
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+ PHGH+TS+AV R +R+LGLA KLM + RAM + F
Sbjct 63 DPDDV--PHGHITSLAVKRSHRRLGLAQKLMDQASRAMIESF 102
> hsa:8260 NAA10, ARD1, ARD1A, DXS707, MGC71248, TE2; N(alpha)-acetyltransferase
10, NatA catalytic subunit (EC:2.3.1.88);
K00670 peptide alpha-N-acetyltransferase [EC:2.3.1.88]
Length=235
Score = 119 bits (299), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 56/102 (54%), Positives = 73/102 (71%), Gaps = 2/102 (1%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+R + DL MQH N + LPENY MKY+F+H LSWPQL +A+D GK+VGYVLAK+E+
Sbjct 3 IRNARPEDLMNMQHCNLLCLPENYQMKYYFYHGLSWPQLSYIAEDENGKIVGYVLAKMEE 62
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+ PHGH+TS+AV R +R+LGLA KLM + RAM + F
Sbjct 63 DPDDV--PHGHITSLAVKRSHRRLGLAQKLMDQASRAMIENF 102
> mmu:97243 Naa11, Ard1b, Ard2, C80008; N(alpha)-acetyltransferase
11, NatA catalytic subunit (EC:2.3.1.88); K00670 peptide
alpha-N-acetyltransferase [EC:2.3.1.88]
Length=218
Score = 119 bits (299), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 57/102 (55%), Positives = 72/102 (70%), Gaps = 5/102 (4%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
RP D L MQH N + LPENY MKY+F+H LSWPQL +A+D GK+VGYVLAK+E+
Sbjct 6 ARPDD---LMNMQHCNLLCLPENYQMKYYFYHGLSWPQLSYIAEDEDGKIVGYVLAKMEE 62
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+ PHGH+TS+AV R +R+LGLA KLM + RAM + F
Sbjct 63 DPDDV--PHGHITSLAVKRSHRRLGLAQKLMDQASRAMIENF 102
> hsa:84779 NAA11, ARD1B, ARD2, MGC10646, hARD2; N(alpha)-acetyltransferase
11, NatA catalytic subunit (EC:2.3.1.88); K00670
peptide alpha-N-acetyltransferase [EC:2.3.1.88]
Length=229
Score = 119 bits (298), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 56/102 (54%), Positives = 72/102 (70%), Gaps = 2/102 (1%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+R + DL MQH N + LPENY MKY+ +H LSWPQL +A+D GK+VGYVLAK+E+
Sbjct 3 IRNAQPDDLMNMQHCNLLCLPENYQMKYYLYHGLSWPQLSYIAEDEDGKIVGYVLAKMEE 62
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
E PHGH+TS+AV R +R+LGLA KLM + RAM + F
Sbjct 63 EPDDV--PHGHITSLAVKRSHRRLGLAQKLMDQASRAMIENF 102
> dre:406643 naa10, ard1a, hm:zeh0223, wu:fc66b08, zgc:63981;
N(alpha)-acetyltransferase 10, NatA catalytic subunit (EC:2.3.1.88)
Length=224
Score = 119 bits (298), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 56/102 (54%), Positives = 73/102 (71%), Gaps = 2/102 (1%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+R + DL MQH N + LPENY MKY+F+H LSWPQL +A+D GK+VGYVLAK+E+
Sbjct 3 IRNARPEDLMNMQHCNLLCLPENYQMKYYFYHGLSWPQLSYIAEDENGKIVGYVLAKMEE 62
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+ PHGH+TS+AV R +R+LGLA KLM + RAM + F
Sbjct 63 DPDDV--PHGHITSLAVKRSHRRLGLAQKLMDQASRAMIENF 102
> dre:100331779 alpha-N-acetyltransferase 1A-like; K00670 peptide
alpha-N-acetyltransferase [EC:2.3.1.88]
Length=224
Score = 119 bits (298), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 56/102 (54%), Positives = 73/102 (71%), Gaps = 2/102 (1%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+R + DL MQH N + LPENY MKY+F+H LSWPQL +A+D GK+VGYVLAK+E+
Sbjct 3 IRNARPEDLMNMQHCNLLCLPENYQMKYYFYHGLSWPQLSYIAEDENGKIVGYVLAKMEE 62
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+ PHGH+TS+AV R +R+LGLA KLM + RAM + F
Sbjct 63 DPDDV--PHGHITSLAVKRSHRRLGLAQKLMDQASRAMIENF 102
> mmu:56292 Naa10, 2310039H09Rik, Ard1, Ard1a, Te2; N(alpha)-acetyltransferase
10, NatA catalytic subunitNalpha acetyltransferase
10 (EC:2.3.1.88); K00670 peptide alpha-N-acetyltransferase
[EC:2.3.1.88]
Length=225
Score = 118 bits (296), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 56/102 (54%), Positives = 73/102 (71%), Gaps = 2/102 (1%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+R + DL MQH N + LPENY MKY+F+H LSWPQL +A+D GK+VGYVLAK+E+
Sbjct 3 IRNARPEDLMNMQHCNLLCLPENYQMKYYFYHGLSWPQLSYIAEDENGKIVGYVLAKMEE 62
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+ PHGH+TS+AV R +R+LGLA KLM + RAM + F
Sbjct 63 DPDDV--PHGHITSLAVKRSHRRLGLAQKLMDQASRAMIENF 102
> cel:K07H8.3 hypothetical protein; K00670 peptide alpha-N-acetyltransferase
[EC:2.3.1.88]
Length=182
Score = 118 bits (295), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 53/102 (51%), Positives = 77/102 (75%), Gaps = 2/102 (1%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+R + V DL MQ+ N + LPENY MKY+F+H+LSWPQL +A+D +G +VGYVLAK+E+
Sbjct 3 IRCARVDDLMSMQNANLMCLPENYQMKYYFYHALSWPQLSYIAEDHKGNVVGYVLAKMEE 62
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+ ++PHGH+TS+AV R R+LGLA+K+M + RAM + +
Sbjct 63 D--PGEEPHGHITSLAVKRSYRRLGLANKMMDQTARAMVETY 102
> cpv:cgd5_3090 N-acetyltransferase subunit ARD1 ; K00670 peptide
alpha-N-acetyltransferase [EC:2.3.1.88]
Length=161
Score = 102 bits (253), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 46/106 (43%), Positives = 69/106 (65%), Gaps = 5/106 (4%)
Query 26 SLVRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQ--GKLVGYVLA 83
+ +R + + DLF +Q N LPENY +KY+++HS++WPQL +A D K VGYVL
Sbjct 2 ACIRRATIDDLFTIQQNNLYCLPENYQIKYYYYHSMTWPQLLDIATDSNDPNKSVGYVLG 61
Query 84 KLEDENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
K+ED++ HGH+TS+AV+R +R GLA KL+ + MQ ++
Sbjct 62 KIEDDSNPL---HGHITSIAVLRTHRGFGLAKKLLTQNHHGMQSIY 104
> pfa:PF10_0036 N-acetyltransferase, putative; K00670 peptide
alpha-N-acetyltransferase [EC:2.3.1.88]
Length=152
Score = 101 bits (251), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 44/102 (43%), Positives = 70/102 (68%), Gaps = 4/102 (3%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+R +++DL MQ N +NLPENY ++Y+F+H+LSWP L +A+D GK+ GY L KLE+
Sbjct 4 IRKCNIYDLLSMQQCNSINLPENYNLRYYFYHALSWPYLSQIAEDVNGKVCGYSLGKLEE 63
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+N + GH+TSVAV++ RKL LA L+ + + ++ ++
Sbjct 64 DN----EYKGHLTSVAVLKTYRKLKLAFYLILQTHQHLKDIY 101
> tpv:TP04_0161 N-acetyltransferase (EC:2.3.1.-); K00670 peptide
alpha-N-acetyltransferase [EC:2.3.1.88]
Length=153
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 41/101 (40%), Positives = 68/101 (67%), Gaps = 4/101 (3%)
Query 29 RPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLEDE 88
R ++++DL + N VN+ ENY MKY+F+H LSWP L + + +G++ GY ++KLE++
Sbjct 5 RRANIYDLVSLSDCNLVNVIENYQMKYYFYHLLSWPHLTNITTNNRGRVCGYSMSKLEED 64
Query 89 NRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+ GHVT+V V+R R LG+A+K+++ + AM KV+
Sbjct 65 ----KNKSGHVTAVGVLRSFRNLGIATKVIKQTHNAMNKVY 101
> bbo:BBOV_II004010 18.m06331; N-acetyltransferase; K00670 peptide
alpha-N-acetyltransferase [EC:2.3.1.88]
Length=172
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 39/102 (38%), Positives = 63/102 (61%), Gaps = 4/102 (3%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLED 87
+R + ++DL N VN+ ENY MKY+F+H LSWPQL +A G + GY +AKLE+
Sbjct 4 IRRASMYDLIGTSDCNLVNVIENYQMKYYFYHLLSWPQLTNIAVSPSGYVCGYSMAKLEE 63
Query 88 ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
+ + GH+T+V V+R R +G+A +++ + AM ++
Sbjct 64 D----VENAGHLTAVGVLRSYRYMGIAKNVIKQTHNAMNAIY 101
> sce:YHR013C ARD1, NAA10; Ard1p (EC:2.3.1.88); K00670 peptide
alpha-N-acetyltransferase [EC:2.3.1.88]
Length=238
Score = 84.7 bits (208), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 49/140 (35%), Positives = 75/140 (53%), Gaps = 39/140 (27%)
Query 28 VRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVA----------------- 70
+R + + D+ MQ+ N NLPENY+MKY+ +H LSWP+ VA
Sbjct 5 IRRATINDIICMQNANLHNLPENYMMKYYMYHILSWPEASFVATTTTLDCEDSDEQDEND 64
Query 71 ---------QDGQG------------KLVGYVLAKLEDE-NRQFQKPHGHVTSVAVMRQN 108
DG+ KLVGYVL K+ D+ ++Q + P+GH+TS++VMR
Sbjct 65 KLELTLDGTNDGRTIKLDPTYLAPGEKLVGYVLVKMNDDPDQQNEPPNGHITSLSVMRTY 124
Query 109 RKLGLASKLMRLSQRAMQKV 128
R++G+A LMR + A+++V
Sbjct 125 RRMGIAENLMRQALFALREV 144
> cpv:cgd1_2660 n-terminal acetyltransferase complex ard1 ; K00670
peptide alpha-N-acetyltransferase [EC:2.3.1.88]
Length=180
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/102 (32%), Positives = 60/102 (58%), Gaps = 3/102 (2%)
Query 19 RLPAKMCSLVRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLV 78
+P + + RP + D+F++ +N + E Y M Y+ + +WP+L V + +
Sbjct 21 NMPQETALIYRPLKLSDIFKINKVNLDSFTETYNMNYYGDYLSTWPELCFVCEAPDHSIA 80
Query 79 GYVLAKLEDENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRL 120
GY+++K+E E Q+ HGHVT+++V +Q R G+A+KLM+
Sbjct 81 GYLVSKVEGEGDQW---HGHVTALSVSQQYRNSGVATKLMKF 119
> tgo:TGME49_089900 N-acetyltransferase 5, putative (EC:2.3.1.88);
K00670 peptide alpha-N-acetyltransferase [EC:2.3.1.88]
Length=166
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 54/104 (51%), Gaps = 3/104 (2%)
Query 26 SLVRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKL 85
S RP DLF +N E ++ ++ + +WP+L VA G + GY++ K+
Sbjct 2 SCFRPMTAGDLFRFNAVNLDGFTETFLQSFYLRYLSNWPELCIVADAPDGTVAGYIIGKV 61
Query 86 EDENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
E + + HGHVT+++V + RK GLA KLM + +K F
Sbjct 62 EGKGMDW---HGHVTALSVAPEFRKCGLARKLMSFLEDISEKKF 102
> cel:Y97E10AL.3 hypothetical protein; K00670 peptide alpha-N-acetyltransferase
[EC:2.3.1.88]
Length=173
Score = 64.7 bits (156), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 30/101 (29%), Positives = 60/101 (59%), Gaps = 3/101 (2%)
Query 26 SLVRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKL 85
+ +RP DV D+F+ ++N E Y +++ + +++P+ VA+ G+++ YV+ K+
Sbjct 2 TTLRPFDVMDMFKFNNVNLDINTETYGFQFYLHYMMNYPEYYQVAEHPNGEIMAYVMGKI 61
Query 86 EDENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQ 126
E + + HGHVT+++V R+LGLA+ +M +R +
Sbjct 62 EGRDTNW---HGHVTALSVAPNFRRLGLAAYMMEFLERTSE 99
> dre:100334419 N-acetyltransferase 5-like
Length=103
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/103 (31%), Positives = 61/103 (59%), Gaps = 1/103 (0%)
Query 26 SLVRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKL 85
+ +R DLF+ ++N L E Y + ++ + WP+ VA+ G+L+GY++ K
Sbjct 2 TTLRAFTCDDLFKFNNINLDPLTETYGIPFYLQYLAHWPEYFIVAEAPGGELMGYIMGKA 61
Query 86 EDENRQFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKV 128
E + ++ HGHVT+++V + R+LGLA+KLM + + +++
Sbjct 62 EGSVAR-EEWHGHVTALSVAPEFRRLGLAAKLMEMLEEISERL 103
> mmu:67877 Naa20, 1500004D14Rik, 2900026I01Rik, AU041458, D2Ertd186e,
MGC144325, Nat5; N(alpha)-acetyltransferase 20, NatB
catalytic subunit (EC:2.3.1.88); K00670 peptide alpha-N-acetyltransferase
[EC:2.3.1.88]
Length=178
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 56/93 (60%), Gaps = 1/93 (1%)
Query 35 DLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLEDENRQFQK 94
DLF ++N L E Y + ++ + WP+ VA+ G+L+GY++ K E + ++
Sbjct 11 DLFRFNNINLDPLTETYGIPFYLQYLAHWPEYFIVAEAPGGELMGYIMGKAEGSVAR-EE 69
Query 95 PHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQK 127
HGHVT+++V + R+LGLA+KLM L + ++
Sbjct 70 WHGHVTALSVAPEFRRLGLAAKLMELLEEISER 102
> hsa:51126 NAA20, NAT3, NAT5, dJ1002M8.1; N(alpha)-acetyltransferase
20, NatB catalytic subunit (EC:2.3.1.88); K00670 peptide
alpha-N-acetyltransferase [EC:2.3.1.88]
Length=178
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 56/93 (60%), Gaps = 1/93 (1%)
Query 35 DLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLEDENRQFQK 94
DLF ++N L E Y + ++ + WP+ VA+ G+L+GY++ K E + ++
Sbjct 11 DLFRFNNINLDPLTETYGIPFYLQYLAHWPEYFIVAEAPGGELMGYIMGKAEGSVAR-EE 69
Query 95 PHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQK 127
HGHVT+++V + R+LGLA+KLM L + ++
Sbjct 70 WHGHVTALSVAPEFRRLGLAAKLMELLEEISER 102
> xla:380338 naa20, MGC53877, nat3, nat5; N(alpha)-acetyltransferase
20, NatB catalytic subunit (EC:2.3.1.88); K00670 peptide
alpha-N-acetyltransferase [EC:2.3.1.88]
Length=178
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 56/93 (60%), Gaps = 1/93 (1%)
Query 35 DLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLEDENRQFQK 94
DLF ++N L E Y + ++ + WP+ VA+ G+L+GY++ K E + ++
Sbjct 11 DLFRFNNINLDPLTETYGIPFYLQYLAHWPEYFIVAEAPGGELMGYIMGKAEGSVAR-EE 69
Query 95 PHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQK 127
HGHVT+++V + R+LGLA+KLM L + ++
Sbjct 70 WHGHVTALSVAPEFRRLGLAAKLMELLEEISER 102
> dre:541516 naa20, im:7150897, nat5, zgc:110819; N(alpha)-acetyltransferase
20, NatB catalytic subunit (EC:2.3.1.88); K00670
peptide alpha-N-acetyltransferase [EC:2.3.1.88]
Length=178
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 57/93 (61%), Gaps = 1/93 (1%)
Query 35 DLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLEDENRQFQK 94
DLF+ ++N L E Y + ++ + WP+ VA+ G+L+GY++ K E + ++
Sbjct 11 DLFKFNNINLDPLTETYGIPFYLQYLAHWPEYFIVAEAPGGELMGYIMGKAEGSVAR-EE 69
Query 95 PHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQK 127
HGHVT+++V + R+LGLA+KLM + + ++
Sbjct 70 WHGHVTALSVAPEFRRLGLAAKLMEMLEEISER 102
> ath:AT1G03150 GCN5-related N-acetyltransferase (GNAT) family
protein; K00670 peptide alpha-N-acetyltransferase [EC:2.3.1.88]
Length=174
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 49/94 (52%), Gaps = 3/94 (3%)
Query 35 DLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLEDENRQFQK 94
DL +N +L E + M ++ + WP VA+ +++GY++ K+E + +
Sbjct 11 DLLRFTSVNLDHLTETFNMSFYMTYLARWPDYFHVAEGPGNRVMGYIMGKVEGQGESW-- 68
Query 95 PHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKV 128
HGHVT+V V + R+ LA KLM L + K+
Sbjct 69 -HGHVTAVTVSPEYRRQQLAKKLMNLLEDISDKI 101
> sce:YPR131C NAT3, NAA20; Catalytic subunit of the NatB N-terminal
acetyltransferase, which catalyzes acetylation of the
amino-terminal methionine residues of all proteins beginning
with Met-Asp or Met-Glu and of some proteins beginning with
Met-Asn or Met-Met (EC:2.3.1.88); K00670 peptide alpha-N-acetyltransferase
[EC:2.3.1.88]
Length=195
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 56/98 (57%), Gaps = 9/98 (9%)
Query 26 SLVRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQL------PTVAQDGQGKLVG 79
+ ++P + DLF+ ++N L EN+ ++++F + + WP L TV + + G
Sbjct 2 TTIQPFEPVDLFKTNNVNLDILTENFPLEFYFEYMIIWPDLFFKSSEMTVDPTFKHNISG 61
Query 80 YVLAKLEDENRQFQKPHGHVTSVAVMRQNRKLGLASKL 117
Y++AK E + ++ H H+T+V V + R++ LASKL
Sbjct 62 YMMAKTEGKTTEW---HTHITAVTVAPRFRRISLASKL 96
> pfa:PFA_0465c N-terminal acetyltransferase, putative (EC:2.3.1.1);
K00670 peptide alpha-N-acetyltransferase [EC:2.3.1.88]
Length=155
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/92 (23%), Positives = 46/92 (50%), Gaps = 3/92 (3%)
Query 35 DLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLEDENRQFQK 94
DL+++ ++N E + K++ + WP + + ++ GY+L K E + +
Sbjct 11 DLYKVNNVNLDPFTEVFNDKFYLRYIYKWPHMNIITKEIDDHTSGYILGKEEGFGQDY-- 68
Query 95 PHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQ 126
HGHVT++++ +R+ G LM ++ Q
Sbjct 69 -HGHVTALSIEEDSRRTGKGVDLMTEFEKITQ 99
> ath:AT2G38130 ATMAK3; ATMAK3; N-acetyltransferase; K00670 peptide
alpha-N-acetyltransferase [EC:2.3.1.88]
Length=190
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 45/82 (54%), Gaps = 4/82 (4%)
Query 46 NLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLEDENRQFQKPHGHVTSVAVM 105
L E Y + + + WPQL +A +GK VG ++ K+ D + F+ G++ + V+
Sbjct 37 ELSEPYSIFTYRYFVYLWPQLCFLAFH-KGKCVGTIVCKMGDHRQTFR---GYIAMLVVI 92
Query 106 RQNRKLGLASKLMRLSQRAMQK 127
+ R G+AS+L+ + +AM +
Sbjct 93 KPYRGRGIASELVTRAIKAMME 114
> cel:B0238.10 hypothetical protein; K00670 peptide alpha-N-acetyltransferase
[EC:2.3.1.88]
Length=278
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 41/77 (53%), Gaps = 6/77 (7%)
Query 51 YVMKYFFFHSLSWPQLPTVAQD-GQGKLVGYVLAKLEDENRQFQKPHGHVTSVAVMRQNR 109
Y +YF + WP+ +A D +G VL KLE + + + G++ +AV R
Sbjct 124 YTYRYFLHN---WPEYCFLAYDQTNNTYIGAVLCKLELD--MYGRCKGYLAMLAVDESCR 178
Query 110 KLGLASKLMRLSQRAMQ 126
+LG+ ++L+R + AMQ
Sbjct 179 RLGIGTRLVRRALDAMQ 195
> pfa:MAL8P1.200 Acetyltransferase, putative
Length=225
Score = 34.7 bits (78), Expect = 0.067, Method: Compositional matrix adjust.
Identities = 31/112 (27%), Positives = 50/112 (44%), Gaps = 16/112 (14%)
Query 34 FDLFEMQHLNFVNLPENYV--MKYFFFHSLSWPQ----LPTVAQDG---------QGKLV 78
FD E+ F P+NY+ M LS P L TV D + + V
Sbjct 66 FDEKEIDIYQFRTFPKNYLNSMYNLLSTELSEPYNIFLLKTVLNDYGEIALMCIFEEQCV 125
Query 79 GYVLAKLEDENRQFQK-PHGHVTSVAVMRQNRKLGLASKLMRLSQRAMQKVF 129
G V++K+ + + + G++ +AV + R LGL S L+ S + MQ ++
Sbjct 126 GAVISKITTKCKNDETITFGYICMIAVHKSIRSLGLGSYLLNESIKLMQNIY 177
> sce:YPR051W MAK3, NAA30; Catalytic subunit of N-terminal acetyltransferase
of the NatC type; required for replication of
dsRNA virus (EC:2.3.1.88); K00670 peptide alpha-N-acetyltransferase
[EC:2.3.1.88]
Length=176
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 41/81 (50%), Gaps = 9/81 (11%)
Query 51 YVMKYFFFHSLSWPQLPTVAQDGQGKL----VGYVLAKLEDENRQFQKPHGHVTSVAVMR 106
YV +YF WP+L +A D + +G ++ K+ D +R + G++ +AV
Sbjct 34 YVYRYFLNQ---WPELTYIAVDNKSGTPNIPIGCIVCKM-DPHRNV-RLRGYIGMLAVES 88
Query 107 QNRKLGLASKLMRLSQRAMQK 127
R G+A KL+ ++ MQ+
Sbjct 89 TYRGHGIAKKLVEIAIDKMQR 109
> xla:100335046 aanat, aanat-b, aanat1, nat4, snat; arylalkylamine
N-acetyltransferase (EC:2.3.1.87)
Length=201
Score = 33.1 bits (74), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 55/111 (49%), Gaps = 14/111 (12%)
Query 20 LPAKMCSLVRPSDVFDLFEMQHLNFVNLPEN---YVMKYFFFHSLSWPQLPTVAQDGQGK 76
LPA + P D +FE++ F+++ ++ + F +L P+L ++ +G+
Sbjct 26 LPASEFRCLSPEDAVSVFEIEREAFISVSGECPLHLDEVRQFLTLC-PEL-SLGWFEEGR 83
Query 77 LVGYVLAKLEDENR------QFQKPHG---HVTSVAVMRQNRKLGLASKLM 118
LV +++ + +++R F KP G H+ +AV R R+ G S L+
Sbjct 84 LVAFIIGSMWNQDRLSQDALTFHKPEGSSVHIHVLAVHRTFRQQGKGSILL 134
> mmu:385377 Pnma5, KIAA1934, mKIAA1934; paraneoplastic antigen
family 5
Length=618
Score = 32.0 bits (71), Expect = 0.53, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 1/32 (3%)
Query 70 AQDGQGKLVGYVLAKLEDENRQFQKPHGHVTS 101
Q GQGK ++LA+ D N+Q Q PH VT+
Sbjct 473 GQQGQGKAPNFLLAR-NDPNKQEQIPHSSVTT 503
> xla:100335045 aanat-a; arylalkylamine N-acetyltransferase a
(EC:2.3.1.87); K00669 arylalkylamine N-acetyltransferase [EC:2.3.1.87]
Length=201
Score = 31.6 bits (70), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 55/111 (49%), Gaps = 14/111 (12%)
Query 20 LPAKMCSLVRPSDVFDLFEMQHLNFVNLPEN---YVMKYFFFHSLSWPQLPTVAQDGQGK 76
LPA + P D +FE++ F+++ ++ + F +L P+L ++ +G+
Sbjct 26 LPASEFRCLSPEDAVSVFEIEREAFISVSGECPLHLDEVRQFLTLC-PEL-SLGWFEEGR 83
Query 77 LVGYVLAKLEDENRQFQ------KPHG---HVTSVAVMRQNRKLGLASKLM 118
LV +++ L +++R Q KP G H+ +AV R R+ G S L+
Sbjct 84 LVAFIIGSLWNQDRLSQDALTLHKPEGSSVHIHVLAVHRTFRQQGKGSILL 134
> cel:F25C8.3 unc-80; UNCoordinated family member (unc-80)
Length=3225
Score = 31.2 bits (69), Expect = 0.89, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 45 VNLPENYVMKYFFFHSLSWPQLPTV 69
++LP YV Y+FFH +PQL V
Sbjct 2153 IHLPAAYVRDYYFFHRSFYPQLTLV 2177
> cel:T12E12.2 hypothetical protein
Length=891
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 26/43 (60%), Gaps = 2/43 (4%)
Query 83 AKLEDENR--QFQKPHGHVTSVAVMRQNRKLGLASKLMRLSQR 123
AKLE++NR Q + PH + +++ + G+ S++ ++S R
Sbjct 133 AKLEEQNRKEQAKNPHASAETEPAVKKKKIEGVGSRIPKISDR 175
> mmu:30805 Slc22a4, Octn1; solute carrier family 22 (organic
cation transporter), member 4; K08201 MFS transporter, OCT family,
solute carrier family 22 (organic cation transporter),
member 4
Length=553
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 22 AKMCSLVRPSDVFDLFEMQHLNFVNLPENYVMKYF 56
AKM S+V P+ +FD E+Q LN + + ++ F
Sbjct 301 AKMNSIVAPAGIFDPLELQELNSLKQQKVIILDLF 335
> tgo:TGME49_033320 hypothetical protein
Length=745
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 24/94 (25%), Positives = 42/94 (44%), Gaps = 3/94 (3%)
Query 27 LVRPSDVFDLFEMQHLNFVNLPENYVMKYFFFHSLSWPQLPTVAQDGQGKLVGYVLAKLE 86
L + S +F LF+++ ++ E ++M YF F L+ + G + VL+ +
Sbjct 147 LTKLSLIFSLFDVKGEGSLSEAEFFLMCYFVFRGLAKAVKRPIP--GYASIEPLVLSTFD 204
Query 87 D-ENRQFQKPHGHVTSVAVMRQNRKLGLASKLMR 119
+ +N Q HG V S V N + S L +
Sbjct 205 ELQNAQCGTKHGSVISNVVGGHNEEADTKSDLQK 238
> ath:AT1G55150 DEAD box RNA helicase, putative (RH20); K12823
ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=501
Score = 27.7 bits (60), Expect = 10.0, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 26/61 (42%), Gaps = 6/61 (9%)
Query 59 HSLSWP------QLPTVAQDGQGKLVGYVLAKLEDENRQFQKPHGHVTSVAVMRQNRKLG 112
S WP L +A+ G GK + Y+L + N Q HG V V+ R+L
Sbjct 126 QSQGWPMAMKGRDLIGIAETGSGKTLSYLLPAIVHVNAQPMLAHGDGPIVLVLAPTRELA 185
Query 113 L 113
+
Sbjct 186 V 186
Lambda K H
0.329 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044474180
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40