bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2105_orf2
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
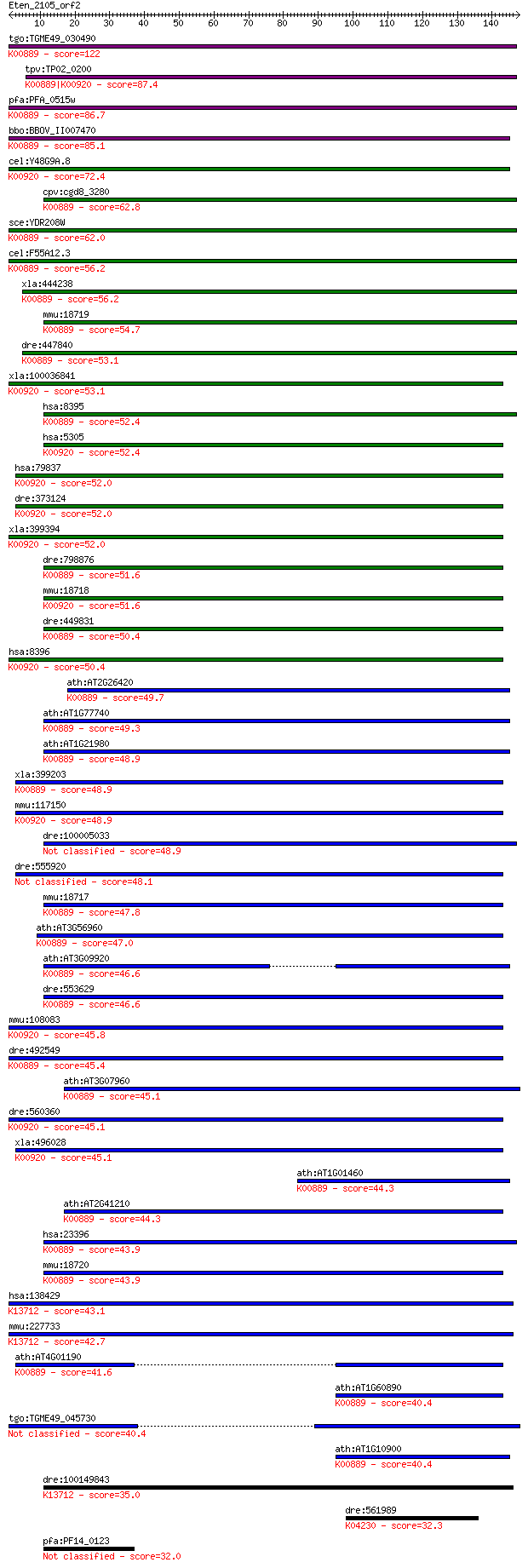
tgo:TGME49_030490 phosphatidylinositol-4-phosphate 5-kinase, p... 122 3e-28
tpv:TP02_0200 phosphatidylinositol-4-phosphate 5-kinase; K0088... 87.4 1e-17
pfa:PFA_0515w PfPIP5K, NCS; phosphatidylinositol-4-phosphate-5... 86.7 3e-17
bbo:BBOV_II007470 18.m06619; 1-phosphatidylinositol-4-phosphat... 85.1 7e-17
cel:Y48G9A.8 ppk-2; PIP Kinase family member (ppk-2); K00920 1... 72.4 5e-13
cpv:cgd8_3280 phosphatidylinositol-4-phosphate 5-kinase ; K008... 62.8 4e-10
sce:YDR208W MSS4; Mss4p (EC:2.7.1.68); K00889 1-phosphatidylin... 62.0 7e-10
cel:F55A12.3 ppk-1; PIP Kinase family member (ppk-1); K00889 1... 56.2 3e-08
xla:444238 pip5k1b, MGC80809; phosphatidylinositol-4-phosphate... 56.2 3e-08
mmu:18719 Pip5k1b, AI844522, Pip5k1a, Pipk5a, Pipk5b, STM7; ph... 54.7 1e-07
dre:447840 fj29b05, fj93b05, wu:fj29b05, wu:fj93b05; zgc:92316... 53.1 3e-07
xla:100036841 pip4k2a, pi5p4ka, pip5k2a, pip5kiia, pipk; phosp... 53.1 3e-07
hsa:8395 PIP5K1B, MSS4, STM7; phosphatidylinositol-4-phosphate... 52.4 5e-07
hsa:5305 PIP4K2A, FLJ13267, PI5P4KA, PIP5K2A, PIP5KII-alpha, P... 52.4 5e-07
hsa:79837 PIP4K2C, FLJ22055, PIP5K2C; phosphatidylinositol-5-p... 52.0 6e-07
dre:373124 pip5k2, Pip5k2c, cb770, pip4k2c, zgc:56046; phospha... 52.0 6e-07
xla:399394 pip4k2b, MGC81828, PI5P4KB, PIP5KIIB, PIP5KIIbeta, ... 52.0 7e-07
dre:798876 si:ch211-197g15.13; K00889 1-phosphatidylinositol-4... 51.6 8e-07
mmu:18718 Pip4k2a, AW742916, Pip5k2a; phosphatidylinositol-5-p... 51.6 9e-07
dre:449831 pip5k1b, zgc:101046; phosphatidylinositol-4-phospha... 50.4 2e-06
hsa:8396 PIP4K2B, PI5P4KB, PIP5K2B, PIP5KIIB, PIP5KIIbeta; pho... 50.4 2e-06
ath:AT2G26420 PIP5K3; PIP5K3 (1-PHOSPHATIDYLINOSITOL-4-PHOSPHA... 49.7 3e-06
ath:AT1G77740 1-phosphatidylinositol-4-phosphate 5-kinase, put... 49.3 5e-06
ath:AT1G21980 ATPIP5K1 (PHOSPHATIDYLINOSITOL-4-PHOSPHATE 5-KIN... 48.9 5e-06
xla:399203 pip5k1a, pip5k1c; phosphatidylinositol-4-phosphate ... 48.9 5e-06
mmu:117150 Pip4k2c, Pip5k2c; phosphatidylinositol-5-phosphate ... 48.9 6e-06
dre:100005033 si:ch211-243a15.1 48.9 6e-06
dre:555920 novel protein similar to vertebrate phosphatidylino... 48.1 8e-06
mmu:18717 Pip5k1c, AI115456, AI835305, Pip5kIgamma; phosphatid... 47.8 1e-05
ath:AT3G56960 PIP5K4; PIP5K4 (PHOSPHATIDYL INOSITOL MONOPHOSPH... 47.0 2e-05
ath:AT3G09920 PIP5K9; PIP5K9 (PHOSPHATIDYL INOSITOL MONOPHOSPH... 46.6 2e-05
dre:553629 pip5k1a, MGC110576, zgc:110576; phosphatidylinosito... 46.6 3e-05
mmu:108083 Pip4k2b, AI848124, PI5P4Kbeta, Pip5k2b, c11; phosph... 45.8 5e-05
dre:492549 im:7140933; zgc:158838; K00889 1-phosphatidylinosit... 45.4 6e-05
ath:AT3G07960 phosphatidylinositol-4-phosphate 5-kinase family... 45.1 8e-05
dre:560360 similar to type II phosphatidylinositolphosphate ki... 45.1 8e-05
xla:496028 pip4k2c, pip5k2c; phosphatidylinositol-5-phosphate ... 45.1 8e-05
ath:AT1G01460 PIPK11; PIPK11; 1-phosphatidylinositol-4-phospha... 44.3 1e-04
ath:AT2G41210 PIP5K5; PIP5K5 (PHOSPHATIDYLINOSITOL- 4-PHOSPHAT... 44.3 1e-04
hsa:23396 PIP5K1C, KIAA0589, LCCS3, PIP5K-GAMMA, PIP5Kgamma, P... 43.9 2e-04
mmu:18720 Pip5k1a, Pip5k1b, Pipk5a, Pipk5b; phosphatidylinosit... 43.9 2e-04
hsa:138429 PIP5KL1, MGC46424, PIPKH, bA203J24.5; phosphatidyli... 43.1 3e-04
mmu:227733 Pip5kl1, BC028795; phosphatidylinositol-4-phosphate... 42.7 4e-04
ath:AT4G01190 ATPIPK10 (Arabidopsis phosphatidylinositol phosp... 41.6 0.001
ath:AT1G60890 phosphatidylinositol-4-phosphate 5-kinase family... 40.4 0.002
tgo:TGME49_045730 phosphatidylinositol-4-phosphate 5-Kinase, p... 40.4 0.002
ath:AT1G10900 phosphatidylinositol-4-phosphate 5-kinase family... 40.4 0.002
dre:100149843 phosphatidylinositol-4-phosphate 5-kinase, type ... 35.0 0.084
dre:561989 similar to galanin receptor type 1; K04230 galanin ... 32.3 0.53
pfa:PF14_0123 conserved Plasmodium protein, unknown function 32.0 0.63
> tgo:TGME49_030490 phosphatidylinositol-4-phosphate 5-kinase,
putative (EC:1.6.3.1 2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=1313
Score = 122 bits (307), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 66/159 (41%), Positives = 94/159 (59%), Gaps = 15/159 (9%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIYY-------------STSGTAASR 47
+++GPER ++LL ++KD FL H ++DYSLL+GI+Y S++ T
Sbjct 1156 MELGPERRSQLLAQMQKDVEFLRVHQIIDYSLLLGIFYRGQQPDDVLAAMVSSTPTPPPP 1215
Query 48 PGGSFSRVPLPAALTNEPIPSASTDTDVPFWRRDLGGLASRDGSKLYYVGIIDVLTKWGA 107
SF L+ PS + VPF++ + GG+ S D SKLYYVGI+D+LT W
Sbjct 1216 RPLSFHHNFRLHHLSGLSQPS--MEPHVPFFQAEFGGMWSADKSKLYYVGIVDILTCWNT 1273
Query 108 FKRTENAIRVVQTCDSYGISCVNPAFYASRFISFLERHI 146
K+ E+A + VQT D ISCVNP +YA RF++F+ RHI
Sbjct 1274 VKKLEHAFKTVQTGDPKSISCVNPDYYAERFMAFMRRHI 1312
> tpv:TP02_0200 phosphatidylinositol-4-phosphate 5-kinase; K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68];
K00920 1-phosphatidylinositol-5-phosphate 4-kinase [EC:2.7.1.149]
Length=848
Score = 87.4 bits (215), Expect = 1e-17, Method: Composition-based stats.
Identities = 50/141 (35%), Positives = 73/141 (51%), Gaps = 23/141 (16%)
Query 6 ERAARLLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSFSRVPLPAALTNEP 65
E + L++LR D NFL ++DYSLL+GI+Y A S+ ++ E
Sbjct 730 EMSESFLQVLRNDVNFLRDSNILDYSLLLGIHYR----AQSKDNVNWDEA--------EK 777
Query 66 IPSASTDTDVPFWRRDLGGLASRDGSKLYYVGIIDVLTKWGAFKRTENAIRVVQTCDSYG 125
+ + T L S + + +YYVGI+DVLT W KR E+ R QT +G
Sbjct 778 LDPGNPHT-----------LMSDNKTSVYYVGIVDVLTTWNLAKRLEHVWRFFQTRQHHG 826
Query 126 ISCVNPAFYASRFISFLERHI 146
+SCVNP FY++RFI + H+
Sbjct 827 VSCVNPVFYSTRFIQSITNHL 847
> pfa:PFA_0515w PfPIP5K, NCS; phosphatidylinositol-4-phosphate-5-kinase
(EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=1710
Score = 86.7 bits (213), Expect = 3e-17, Method: Composition-based stats.
Identities = 56/147 (38%), Positives = 83/147 (56%), Gaps = 7/147 (4%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIYY-STSGTAASRPGGSFSRVPLPA 59
I+IGPE RLL++L+ DA+FL + L+DYSLL GI+Y S + +++
Sbjct 1569 INIGPENKERLLKVLKADADFLKENMLLDYSLLFGIHYRELSKDVVNWEETKTNQINHIY 1628
Query 60 ALTNEPIPSASTDTDVPFWRRDLGGLASRDGSKLYYVGIIDVLTKWGAFKRTENAIRVVQ 119
I S PF + D GG+ S D K+++ GIID+ TKW K+ E+ R +Q
Sbjct 1629 DYKGNCIASR------PFHQCDYGGIISVDKKKIFFFGIIDIFTKWSIKKKFEHTFRTIQ 1682
Query 120 TCDSYGISCVNPAFYASRFISFLERHI 146
D ISC++P YA RF++F+E H+
Sbjct 1683 KFDGKNISCIHPNAYAKRFVTFIENHM 1709
> bbo:BBOV_II007470 18.m06619; 1-phosphatidylinositol-4-phosphate
5-kinase (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=662
Score = 85.1 bits (209), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 50/149 (33%), Positives = 75/149 (50%), Gaps = 35/149 (23%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSFSRVPLPAA 60
I++G ER++ ++L++D FL LMDYSLL+GI+Y
Sbjct 541 IELGSERSSAFFDVLQRDVAFLSDSMLMDYSLLLGIHYR--------------------- 579
Query 61 LTNEPIPSASTDTDVPFWRRD-----LGGLASRDGSKLYYVGIIDVLTKWGAFKRTENAI 115
S D W +D + + + KLY+VGIIDVLTKW KR E+A
Sbjct 580 ---------SISHDEVDWNKDPDHSQVSCIMGKRRDKLYFVGIIDVLTKWDLRKRAESAW 630
Query 116 RVVQTCDSYGISCVNPAFYASRFISFLER 144
R +QT ++ G+SCV P Y R ++F+++
Sbjct 631 RKLQTMNNQGVSCVPPLQYGERLLAFIKQ 659
> cel:Y48G9A.8 ppk-2; PIP Kinase family member (ppk-2); K00920
1-phosphatidylinositol-5-phosphate 4-kinase [EC:2.7.1.149]
Length=401
Score = 72.4 bits (176), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 52/162 (32%), Positives = 79/162 (48%), Gaps = 23/162 (14%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIY--YSTSGTAASRPGGSFSR---- 54
+++ PE L+E+L D +L LMDYSLLVGI+ + AA+RP S
Sbjct 242 LNLPPEAGKLLIEMLTSDTEWLTRMHLMDYSLLVGIHDCERAAQEAANRPVEQNSEESGD 301
Query 55 --VPLPAALTNEPIP---------SASTDTDVPFWRRDLGGLASRDGSKLYYVGIIDVLT 103
P P + PIP S D D F+ + A + + +Y++G++D+LT
Sbjct 302 ELAPTPP---DSPIPSTGGAFPGVSGGPDLDDEFYA--IASPADFEKNLIYFIGLVDILT 356
Query 104 KWGAFKRTENAIRVVQT-CDSYGISCVNPAFYASRFISFLER 144
+G KR+ A + V+ D+ IS V P YA R + F+ R
Sbjct 357 YYGIKKRSATAAKTVKYGSDAENISTVKPEQYAKRLVEFVSR 398
> cpv:cgd8_3280 phosphatidylinositol-4-phosphate 5-kinase ; K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=828
Score = 62.8 bits (151), Expect = 4e-10, Method: Composition-based stats.
Identities = 39/136 (28%), Positives = 60/136 (44%), Gaps = 24/136 (17%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSFSRVPLPAALTNEPIPSAS 70
LL+ L D F + +MDYSLL+GI+Y + + + + N+
Sbjct 715 LLKQLEVDCKFFELYNIMDYSLLIGIHYVQDDA-----------MNIKSLIENKN----- 758
Query 71 TDTDVPFWRRDLGGLASRDGSKLYYVGIIDVLTKWGAFKRTENAIRVVQTCDSYGISCVN 130
R G+ S D K+YY+G ID + W K+ E + + S GIS V
Sbjct 759 --------NRKFVGILSSDQKKVYYLGFIDFFSTWSTIKKFERLYKAIIIGSSNGISAVP 810
Query 131 PAFYASRFISFLERHI 146
P Y+ RF+ F++ I
Sbjct 811 PDKYSMRFMKFIKSKI 826
> sce:YDR208W MSS4; Mss4p (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=779
Score = 62.0 bits (149), Expect = 7e-10, Method: Composition-based stats.
Identities = 45/151 (29%), Positives = 67/151 (44%), Gaps = 7/151 (4%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSFSRVPLPAA 60
I GP + L L+KD L MDYSLL+GI+ + +
Sbjct 608 IKFGPLKKKTFLTQLKKDVELLAKLNTMDYSLLIGIHDINKAKEDDLQLADTASIE-EQP 666
Query 61 LTNEPIPSASTDTDVPFWRRDLGGLASRDGSK-----LYYVGIIDVLTKWGAFKRTENAI 115
T PI + + F+R GG+ + D +YYVGIID LT + K+ E
Sbjct 667 QTQGPIRTGTGTVVRHFFREFEGGIRASDQFNNDVDLIYYVGIIDFLTNYSVMKKLETFW 726
Query 116 RVVQTCDSYGISCVNPAFYASRFISFLERHI 146
R ++ D+ +S + P YA+RF F+E +
Sbjct 727 RSLRH-DTKLVSAIPPRDYANRFYEFIEDSV 756
> cel:F55A12.3 ppk-1; PIP Kinase family member (ppk-1); K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=611
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 50/173 (28%), Positives = 74/173 (42%), Gaps = 43/173 (24%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSFSRVPLPAA 60
I I P L++ + +D + + +MDYSLLVGI+ G S
Sbjct 297 IFIDPTALEALMKSISRDCLVMESFKIMDYSLLVGIHNVELGIKERAEQNRES------- 349
Query 61 LTNEPIPSASTDT------------------------DVPFWRRDLGGLASR--DGSKL- 93
P PS S+DT DVP GG+ +R +G +L
Sbjct 350 ---HPAPSTSSDTFSGEKAENHKVLQEKFSVWDTGDGDVPH-----GGVPARNSNGDRLV 401
Query 94 YYVGIIDVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFLERHI 146
Y+GIID+L + K+ E+ + + D IS NP FYASRF++F+ +
Sbjct 402 LYLGIIDILQNYRLLKKMEHTWKAILH-DGDTISVHNPNFYASRFLTFMTEKV 453
> xla:444238 pip5k1b, MGC80809; phosphatidylinositol-4-phosphate
5-kinase, type I, beta (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=537
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 46/157 (29%), Positives = 80/157 (50%), Gaps = 20/157 (12%)
Query 5 PERAARLLELLRKDANFLVTHGLMDYSLLVGIYY--STS----GTAA------SRPGGSF 52
PE + L++ L++D L + +MDYSLL+G++ STS G ++ RP S
Sbjct 242 PETYSALMKTLQRDCRVLESFKIMDYSLLLGVHVLGSTSKEKEGESSHYSIDNKRP--SV 299
Query 53 SRVPLPAALTNEPIPSASTDTDVPFWRRDLGGLASRD--GSK-LYYVGIIDVLTKWGAFK 109
+V AL E I D + +GG+ +++ G + L ++GIID+L + K
Sbjct 300 QKVLYSTAL--ESIQGPGKDVNSFIKESTMGGIPAKNHKGERMLLFMGIIDILQSYRLMK 357
Query 110 RTENAIRVVQTCDSYGISCVNPAFYASRFISFLERHI 146
+ E++ + + D +S P+FYA RF+ F+ +
Sbjct 358 KLEHSWKAL-VYDGDSVSVHRPSFYADRFLKFMNSRV 393
> mmu:18719 Pip5k1b, AI844522, Pip5k1a, Pipk5a, Pipk5b, STM7;
phosphatidylinositol-4-phosphate 5-kinase, type 1 beta (EC:2.7.1.68);
K00889 1-phosphatidylinositol-4-phosphate 5-kinase
[EC:2.7.1.68]
Length=539
Score = 54.7 bits (130), Expect = 1e-07, Method: Composition-based stats.
Identities = 43/151 (28%), Positives = 71/151 (47%), Gaps = 18/151 (11%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTA------------ASRPGGSFSRVPLP 58
L++ L++D L + +MDYSLL+GI+ A RPG +V
Sbjct 248 LMKTLQRDCRVLESFKIMDYSLLLGIHILDHSLKDKEEEPLQNVPDAKRPG--MQKVLYS 305
Query 59 AALTNEPIPSASTDTDVPFWRRDLGGL--ASRDGSK-LYYVGIIDVLTKWGAFKRTENAI 115
A+ + P S D + +GG+ S G K L ++GIID+L + K+ E++
Sbjct 306 TAMESIQGPGKSADGIIAENPDTMGGIPAKSHKGEKLLLFMGIIDILQSYRLMKKLEHSW 365
Query 116 RVVQTCDSYGISCVNPAFYASRFISFLERHI 146
+ + D +S P+FYA RF+ F+ +
Sbjct 366 KAL-VYDGDTVSVHRPSFYADRFLKFMNSRV 395
> dre:447840 fj29b05, fj93b05, wu:fj29b05, wu:fj93b05; zgc:92316
(EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=527
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 40/151 (26%), Positives = 71/151 (47%), Gaps = 12/151 (7%)
Query 5 PERAARLLELLRKDANFLVTHGLMDYSLLVGIYY------STSGTAASRPGGSFSRVPLP 58
PE L++ L++D L + +MDYSLL+G++ G+A +V
Sbjct 241 PETYNALMKTLQRDCRVLESFKIMDYSLLLGVHVLDQSHRDGDGSAVDGKRTVGQKVLYS 300
Query 59 AALTNEPIPSASTDTDVPFWRRDLGGLAS---RDGSKLYYVGIIDVLTKWGAFKRTENAI 115
A+ E I + +GG+ + RD L ++GIID+L + K+ E++
Sbjct 301 TAM--ESIQGDGKAAEALTTDDTMGGIPAKTHRDEKVLIFLGIIDILQSYRFIKKLEHSW 358
Query 116 RVVQTCDSYGISCVNPAFYASRFISFLERHI 146
+ + D +S P+FYA+RF+ F+ +
Sbjct 359 KAL-VYDGDTVSVHRPSFYANRFLKFMSSRV 388
> xla:100036841 pip4k2a, pi5p4ka, pip5k2a, pip5kiia, pipk; phosphatidylinositol-5-phosphate
4-kinase, type II, alpha (EC:2.7.1.149);
K00920 1-phosphatidylinositol-5-phosphate 4-kinase
[EC:2.7.1.149]
Length=415
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 47/160 (29%), Positives = 71/160 (44%), Gaps = 18/160 (11%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSFSRVPLPAA 60
+ +G E LE L++D FL +MDYSLLVG++ +
Sbjct 251 LHVGEENKKMFLEKLKRDVEFLSVLKIMDYSLLVGLHDVDRAEQEEMEVEERAEEEECED 310
Query 61 LTNEPIPSASTDTDVP-----FWR----------RDLGGLASRDGS---KLYYVGIIDVL 102
+ PI S T D P F R D+ + S D + ++Y++ IID+L
Sbjct 311 GSGNPICSYGTPPDSPGNLLNFPRFFGPGEFDPSVDVYAMKSHDNAPKKEVYFMAIIDIL 370
Query 103 TKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
T + A K+ +A + V+ IS VNP Y+ RFI F+
Sbjct 371 TPYDAKKKAAHAAKTVKHGAGAEISTVNPEQYSKRFIEFM 410
> hsa:8395 PIP5K1B, MSS4, STM7; phosphatidylinositol-4-phosphate
5-kinase, type I, beta (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=540
Score = 52.4 bits (124), Expect = 5e-07, Method: Composition-based stats.
Identities = 39/149 (26%), Positives = 70/149 (46%), Gaps = 14/149 (9%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIYY----------STSGTAASRPGGSFSRVPLPAA 60
L++ L++D L + +MDYSLL+GI++ T +V A
Sbjct 248 LMKTLQRDCRVLESFKIMDYSLLLGIHFLDHSLKEKEEETPQNVPDAKRTGMQKVLYSTA 307
Query 61 LTNEPIPSASTDTDVPFWRRDLGGLASRD--GSK-LYYVGIIDVLTKWGAFKRTENAIRV 117
+ + P S D + +GG+ ++ G K L ++GIID+L + K+ E++ +
Sbjct 308 MESIQGPGKSGDGIITENPDTMGGIPAKSHRGEKLLLFMGIIDILQSYRLMKKLEHSWKA 367
Query 118 VQTCDSYGISCVNPAFYASRFISFLERHI 146
+ D +S P+FYA RF+ F+ +
Sbjct 368 L-VYDGDTVSVHRPSFYADRFLKFMNSRV 395
> hsa:5305 PIP4K2A, FLJ13267, PI5P4KA, PIP5K2A, PIP5KII-alpha,
PIP5KIIA, PIPK; phosphatidylinositol-5-phosphate 4-kinase,
type II, alpha (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=406
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 44/152 (28%), Positives = 70/152 (46%), Gaps = 25/152 (16%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIY-----------------YSTSGTAASRPGGSFS 53
LE L+KD FL LMDYSLLVGI+ + + P G+
Sbjct 255 FLEKLKKDVEFLAQLKLMDYSLLVGIHDVERAEQEEVECEENDGEEEGESDGTHPVGTPP 314
Query 54 RVPLPAALTNEPIPSASTDTDVPFWRRDLGGLASRDGS---KLYYVGIIDVLTKWGAFKR 110
P ++ P+ D ++ D+ G+ + S ++Y++ IID+LT + A K+
Sbjct 315 DSPGNTLNSSPPLAPGEFDPNI-----DVYGIKCHENSPRKEVYFMAIIDILTHYDAKKK 369
Query 111 TENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
+A + V+ IS VNP Y+ RF+ F+
Sbjct 370 AAHAAKTVKHGAGAEISTVNPEQYSKRFLDFI 401
> hsa:79837 PIP4K2C, FLJ22055, PIP5K2C; phosphatidylinositol-5-phosphate
4-kinase, type II, gamma (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=421
Score = 52.0 bits (123), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 50/163 (30%), Positives = 75/163 (46%), Gaps = 23/163 (14%)
Query 3 IGPERAARLLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPG---GSFSRVPLPA 59
IG E LE L++D FLV +MDYSLL+GI+ G+ S V
Sbjct 254 IGEEEKKIFLEKLKRDVEFLVQLKIMDYSLLLGIHDIIRGSEPEEEAPVREDESEVDGDC 313
Query 60 ALTNEPIPSASTDTDVP------FWRRDLG-----------GLASRDGS---KLYYVGII 99
+LT P S T R LG + S +G+ ++Y++G+I
Sbjct 314 SLTGPPALVGSYGTSPEGIGGYIHSHRPLGPGEFESFIDVYAIRSAEGAPQKEVYFMGLI 373
Query 100 DVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
D+LT++ A K+ +A + V+ IS V+P YA RF+ F+
Sbjct 374 DILTQYDAKKKAAHAAKTVKHGAGAEISTVHPEQYAKRFLDFI 416
> dre:373124 pip5k2, Pip5k2c, cb770, pip4k2c, zgc:56046; phosphatidylinositol-4-phosphate
5-kinase, type II (EC:2.7.1.149);
K00920 1-phosphatidylinositol-5-phosphate 4-kinase [EC:2.7.1.149]
Length=416
Score = 52.0 bits (123), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 48/165 (29%), Positives = 78/165 (47%), Gaps = 28/165 (16%)
Query 3 IGPERAARLLELLRKDANFLVTHGLMDYSLLVGIY----------------YS-----TS 41
+ E+ +++E L +D FLV +MDYSLL+GI+ Y +
Sbjct 250 VTEEQKEKMMEKLNRDVEFLVKLKIMDYSLLLGIHDVARGEREEEEAEEPCYEDDADPEN 309
Query 42 GTAASRPGGSFSRVPLP-AALTNEPIPSASTDTDVPFWRRDLGGLASRDGS---KLYYVG 97
G A + GS+ P A N P + D P+ D+ + S G+ ++Y++G
Sbjct 310 GLAPALQVGSYGTSPEGIAGYMNSIKPLGPGEFD-PYI--DVYAVKSAPGAPQREVYFMG 366
Query 98 IIDVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
+IDVLT++ K+ +A + V+ IS V+P YA RF F+
Sbjct 367 LIDVLTQYDTKKKAAHAAKTVKHGAGAEISTVHPEQYAKRFREFI 411
> xla:399394 pip4k2b, MGC81828, PI5P4KB, PIP5KIIB, PIP5KIIbeta,
pip5k2b; phosphatidylinositol-5-phosphate 4-kinase, type II,
beta (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=417
Score = 52.0 bits (123), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 47/160 (29%), Positives = 71/160 (44%), Gaps = 18/160 (11%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSFSRVPLPAA 60
+ +G E LE L++D FL +MDYSLLVG++ +
Sbjct 253 LHVGEENKKIFLEKLKRDVEFLSVLKIMDYSLLVGLHDVERAEQEEMEVEERAEEEECED 312
Query 61 LTNEPIPSASTDTDVP-----FWR----------RDLGGLASRDGS---KLYYVGIIDVL 102
+ PI S T D P F R D+ + S D + ++Y++ IID+L
Sbjct 313 GSGNPICSYGTPPDSPGNLLNFPRFFGPGEFDPSVDVYAMRSHDNAPRKEVYFMAIIDIL 372
Query 103 TKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
T + A K+ +A + V+ IS VNP Y+ RFI F+
Sbjct 373 TPYDAKKKAAHAAKTVKHGAGAEISTVNPEQYSKRFIEFM 412
> dre:798876 si:ch211-197g15.13; K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=682
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 43/148 (29%), Positives = 69/148 (46%), Gaps = 22/148 (14%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASR----PGGSFSRVPLPA----ALT 62
L++ L++D L + +MDYSLL+G++ + G S + PL
Sbjct 303 LVKTLQRDCLVLESFKIMDYSLLLGVHNIDQAAKEQQMEGSQGNSDEKRPLAQKALYTTA 362
Query 63 NEPIPSAST-----DTDVPFWRRDLGGLASRDGSK---LYYVGIIDVLTKWGAFKRTENA 114
E I AS DTD +GG+ + +G L Y+GIID+L + K+ E+
Sbjct 363 MESIQGASACGEGIDTD-----DTMGGIPAVNGRGERLLLYIGIIDILQSYRLIKKLEHT 417
Query 115 IRVVQTCDSYGISCVNPAFYASRFISFL 142
+ + D +S P+FYA RF+ F+
Sbjct 418 WKAL-VHDGDTVSVHRPSFYADRFLRFM 444
> mmu:18718 Pip4k2a, AW742916, Pip5k2a; phosphatidylinositol-5-phosphate
4-kinase, type II, alpha (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=405
Score = 51.6 bits (122), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 43/149 (28%), Positives = 68/149 (45%), Gaps = 19/149 (12%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIY-----------------YSTSGTAASRPGGSFS 53
LE L+KD FL LMDYSLLVGI+ + ++ P G+
Sbjct 255 FLEKLKKDVEFLAQLKLMDYSLLVGIHDVERAEQEEVECEENDGEEEGESDSTHPIGTPP 314
Query 54 RVPLPAALTNEPIPSASTDTDVPFWRRDLGGLASRDGSKLYYVGIIDVLTKWGAFKRTEN 113
P ++ P+ D ++ + A R ++Y++ IID+LT + A K+ +
Sbjct 315 DSPGNTLNSSPPLAPGEFDPNIDVYAIKCHENAPR--KEVYFMAIIDILTHYDAKKKAAH 372
Query 114 AIRVVQTCDSYGISCVNPAFYASRFISFL 142
A + V+ IS VNP Y+ RF+ F+
Sbjct 373 AAKTVKHGAGAEISTVNPEQYSKRFLDFI 401
> dre:449831 pip5k1b, zgc:101046; phosphatidylinositol-4-phosphate
5-kinase, type I, beta (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=521
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 42/157 (26%), Positives = 70/157 (44%), Gaps = 45/157 (28%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIY-----------------------YSTSGTAASR 47
L++ L++D L + +MDYSLL+GI+ YST+ +
Sbjct 247 LMKTLQRDCRVLESFKIMDYSLLLGIHVLDRRIRRGGRGDSRRQGTQKVLYSTARESIQG 306
Query 48 PGGSFSRVPLPAALTNEPIPSASTDTDVPFWRRDLGGLAS--RDGSKLYYVGIIDVLTKW 105
G + P EP+ A +T LGG+ + +D L ++GIID+L +
Sbjct 307 DG----KAP-------EPVADADDET--------LGGIPAKHKDEKLLIFLGIIDILQSY 347
Query 106 GAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
K+ E++ + + D +S P FYA RF+ F+
Sbjct 348 RFIKKVEHSWKAL-VHDGDTVSVHRPNFYADRFLKFM 383
> hsa:8396 PIP4K2B, PI5P4KB, PIP5K2B, PIP5KIIB, PIP5KIIbeta; phosphatidylinositol-5-phosphate
4-kinase, type II, beta (EC:2.7.1.149);
K00920 1-phosphatidylinositol-5-phosphate 4-kinase
[EC:2.7.1.149]
Length=416
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 48/177 (27%), Positives = 70/177 (39%), Gaps = 50/177 (28%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIY----------------------- 37
+ +G E LE L++D FL +MDYSLLVGI+
Sbjct 250 LHVGEESKKNFLEKLKRDVEFLAQLKIMDYSLLVGIHDVDRAEQEEMEVEERAEDEECEN 309
Query 38 -------YSTSGTAASRPGG--SFSRVPLPAALTNEPIPSASTDTDVPFWRRDLGGLASR 88
+ GT PG SF R P PS D+ + S
Sbjct 310 DGVGGNLLCSYGTPPDSPGNLLSFPRFFGPGEFD----PSV-----------DVYAMKSH 354
Query 89 DGS---KLYYVGIIDVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
+ S ++Y++ IID+LT + K+ +A + V+ IS VNP Y+ RF F+
Sbjct 355 ESSPKKEVYFMAIIDILTPYDTKKKAAHAAKTVKHGAGAEISTVNPEQYSKRFNEFM 411
> ath:AT2G26420 PIP5K3; PIP5K3 (1-PHOSPHATIDYLINOSITOL-4-PHOSPHATE
5-KINASE 3); 1-phosphatidylinositol-4-phosphate 5-kinase;
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=705
Score = 49.7 bits (117), Expect = 3e-06, Method: Composition-based stats.
Identities = 37/148 (25%), Positives = 61/148 (41%), Gaps = 26/148 (17%)
Query 18 DANFLVTHGLMDYSLLVGIYYSTSGTAAS---------------RPGGSFSRVPLPAALT 62
D FL +MDYSLL+G+++ SG R G R+
Sbjct 557 DCEFLEAERIMDYSLLIGLHFRESGMRDDISLGIGRRDQEDKLMRGNGPLMRLGESTPAK 616
Query 63 NEPIPSASTDTDVPFWRRDLGGLASRDGSK------LYYVGIIDVLTKWGAFKRTENAIR 116
E + +T W D ++ G++ + Y G+ID+L + K+ E+A +
Sbjct 617 AEQVSRFEEET----WEEDAIDNSNPKGTRKEAVEVILYFGVIDILQDYDITKKLEHAYK 672
Query 117 VVQTCDSYGISCVNPAFYASRFISFLER 144
+ D IS V+P Y+ RF F+ +
Sbjct 673 SLHA-DPASISAVDPKLYSRRFRDFINK 699
> ath:AT1G77740 1-phosphatidylinositol-4-phosphate 5-kinase, putative
/ PIP kinase, putative / PtdIns(4)P-5-kinase, putative
/ diphosphoinositide kinase, putative; K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=754
Score = 49.3 bits (116), Expect = 5e-06, Method: Composition-based stats.
Identities = 44/172 (25%), Positives = 67/172 (38%), Gaps = 39/172 (22%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIYYST---------------SGTAASRPGGSF--- 52
L+ +++D FL +MDYSLLVG+++ SG S F
Sbjct 578 LMTQIKRDCEFLEAERIMDYSLLVGVHFRDDNTGDKMGLSPFVLRSGKIESYQSEKFMRG 637
Query 53 ----------------SRVPLPAALTNEPIPSASTDTDVPFWRRDLGGLASRDGSKLY-- 94
R PL N P + + + GG + ++Y
Sbjct 638 CRFLEAELQDMDRILAGRKPLIRLGANMPARAERMARRSDYDQYSSGGTNYQSHGEVYEV 697
Query 95 --YVGIIDVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFLER 144
Y GIID+L + K+ E+A + +Q D IS V+P Y+ RF F+ R
Sbjct 698 VLYFGIIDILQDYDISKKIEHAYKSLQA-DPASISAVDPKLYSRRFRDFISR 748
> ath:AT1G21980 ATPIP5K1 (PHOSPHATIDYLINOSITOL-4-PHOSPHATE 5-KINASE
1); 1-phosphatidylinositol-4-phosphate 5-kinase/ actin
filament binding / actin monomer binding / phosphatidylinositol
phosphate kinase; K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=752
Score = 48.9 bits (115), Expect = 5e-06, Method: Composition-based stats.
Identities = 41/172 (23%), Positives = 65/172 (37%), Gaps = 39/172 (22%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSF------------------ 52
L++ +++D FL +MDYSLLVG+++ T F
Sbjct 576 LMKQIKRDCEFLEAERIMDYSLLVGVHFRDDNTGEKMGLSPFVLRSGRIDSYQNEKFMRG 635
Query 53 ----------------SRVPLPAALTNEPIPSASTDTDVPFWRRDLGGLASRDGSKLY-- 94
R P N P + F + GG + ++Y
Sbjct 636 CRFLEAELQDMDRILAGRKPSIRLGANMPAKAERMARRSDFDQYSSGGASYPSHGEMYEV 695
Query 95 --YVGIIDVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFLER 144
Y G+ID+L + K+ E+A + +Q D IS V+P Y+ RF F+ R
Sbjct 696 VLYFGVIDILQDYDITKKIEHAYKSLQA-DPASISAVDPKLYSKRFRDFISR 746
> xla:399203 pip5k1a, pip5k1c; phosphatidylinositol-4-phosphate
5-kinase, type I, alpha (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=570
Score = 48.9 bits (115), Expect = 5e-06, Method: Composition-based stats.
Identities = 40/154 (25%), Positives = 69/154 (44%), Gaps = 20/154 (12%)
Query 3 IGPERAARLLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSFSRVPLPAA-- 60
+ P+ L + +++D L + +MDYSLLVG++ + + S PAA
Sbjct 274 LDPDMHNALCKTIQRDCLVLQSFKIMDYSLLVGVHNMDFAQKETMVVDAQSDTRRPAAQK 333
Query 61 ---------LTNEPIPSASTDTDVPFWRRDLGGLASRDGSK---LYYVGIIDVLTKWGAF 108
+ E + +TD +GG+ +R+ L Y+G+IDVL +
Sbjct 334 ALYSTAMESIQGEARRGGAIETD-----DQMGGIPARNAKGERLLLYIGVIDVLQSYRFM 388
Query 109 KRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
K+ E++ + + D +S P FYA RF F+
Sbjct 389 KKLEHSWKAL-VHDGDTVSVHRPGFYADRFQKFM 421
> mmu:117150 Pip4k2c, Pip5k2c; phosphatidylinositol-5-phosphate
4-kinase, type II, gamma (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=421
Score = 48.9 bits (115), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 47/168 (27%), Positives = 79/168 (47%), Gaps = 33/168 (19%)
Query 3 IGPERAARLLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTA------------------ 44
IG E LE L++D FLV +MDYSLL+GI+ G+
Sbjct 254 IGEEEKKVFLEKLKRDVEFLVQLKIMDYSLLLGIHDIIRGSEPEEEGPVREEESEWDGDC 313
Query 45 --ASRPG--GSFSRVPLPAA---LTNEPIPSASTDTDVPFWRRDLGGLASRDGS---KLY 94
A P GS+ P ++ P+ ++ + D+ + S +G+ ++Y
Sbjct 314 NLAGPPALVGSYGTSPEGIGGYIHSHRPLGPGEFESFI-----DVYAIRSAEGAPQKEVY 368
Query 95 YVGIIDVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
++G+ID+LT++ A K+ +A + V+ IS V+P YA RF+ F+
Sbjct 369 FMGLIDILTQYDAKKKAAHAAKTVKHGAGAEISTVHPEQYAKRFLDFI 416
> dre:100005033 si:ch211-243a15.1
Length=802
Score = 48.9 bits (115), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 41/146 (28%), Positives = 66/146 (45%), Gaps = 11/146 (7%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSF-SRVPLPAA------LTN 63
LL+ L++D L + +MDYSLL+G++ + GS S PAA
Sbjct 341 LLKTLQRDCLVLESFKIMDYSLLLGVHNVEQAERERQMEGSQGSDEKRPAAQRALYSTAM 400
Query 64 EPIPSASTDTDVPFWRRDLGGLASRDGSK---LYYVGIIDVLTKWGAFKRTENAIRVVQT 120
E I + D +GG+ + G L ++GIID+L + K+ E+ + +
Sbjct 401 ESIQGGAACGDSIDTDDTMGGIPAVSGKGERLLLFIGIIDILQSYRLIKKLEHTWKAL-V 459
Query 121 CDSYGISCVNPAFYASRFISFLERHI 146
D +S P FYA RF F+ ++
Sbjct 460 HDGDTVSVHRPNFYADRFFRFMSANV 485
> dre:555920 novel protein similar to vertebrate phosphatidylinositol-4-phosphate
5-kinase, type II, alpha (PIP5K2A)
Length=413
Score = 48.1 bits (113), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 45/158 (28%), Positives = 68/158 (43%), Gaps = 18/158 (11%)
Query 3 IGPERAARLLELLRKDANFLVTHGLMDYSLLVGIY-------YSTSGTAASRPGGSFSRV 55
I E LE L+ D FL LMDYSLLVGI+ + G
Sbjct 251 IDDENKKMFLEKLKNDVEFLAQLKLMDYSLLVGIHDVERGEQEQPEEESEDNDAGEEEGT 310
Query 56 PLPAALTNEP--IPSASTDTDVPFWRR------DLGGLASRDGS---KLYYVGIIDVLTK 104
T P PS + D++ P D+ + S + + ++Y++ +ID+LT
Sbjct 311 ESDGGATGSPPDSPSNTLDSNRPLGPGEFDPIIDVYAIKSHENAPKKEIYFMAVIDILTP 370
Query 105 WGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
+ A K+ +A + V+ IS VNP Y+ RF F+
Sbjct 371 YDAKKKAAHAAKTVKHGAGAEISTVNPEQYSKRFYDFI 408
> mmu:18717 Pip5k1c, AI115456, AI835305, Pip5kIgamma; phosphatidylinositol-4-phosphate
5-kinase, type 1 gamma (EC:2.7.1.68);
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=635
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 35/143 (24%), Positives = 68/143 (47%), Gaps = 12/143 (8%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSFSRVPLPAALTNEPIPSAS 70
L++ L++D L + +MDYSLL+G++ + G+ S+ + + + S +
Sbjct 298 LVKTLQRDCLVLESFKIMDYSLLLGVHNIDQQERERQAEGAQSKADEKRPVAQKALYSTA 357
Query 71 TDTDVPFWRR--------DLGGLASRDGSK---LYYVGIIDVLTKWGAFKRTENAIRVVQ 119
++ R +GG+ + +G L ++GIID+L + K+ E+ + +
Sbjct 358 MESIQGGAARGEAIETDDTMGGIPAVNGRGERLLLHIGIIDILQSYRFIKKLEHTWKAL- 416
Query 120 TCDSYGISCVNPAFYASRFISFL 142
D +S P+FYA RF F+
Sbjct 417 VHDGDTVSVHRPSFYAERFFKFM 439
> ath:AT3G56960 PIP5K4; PIP5K4 (PHOSPHATIDYL INOSITOL MONOPHOSPHATE
5 KINASE 4); 1-phosphatidylinositol-4-phosphate 5-kinase;
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=779
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 45/161 (27%), Positives = 65/161 (40%), Gaps = 28/161 (17%)
Query 9 ARLLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTA-------ASRPGGSFSRVPLPA-- 59
+ + KD FL +MDYSLLVGI++ + A A P G F P
Sbjct 612 QEFIRQVDKDCEFLEQERIMDYSLLVGIHFREASVAGELIPSGARTPIGEFEDESAPRLS 671
Query 60 -----ALTNEPIPSAS--------TDTDVPFWRRDLGGLASRDGSKLYY-----VGIIDV 101
L ++P AS + R D + + YY GIID+
Sbjct 672 RADVDQLLSDPTRWASIRLGGNMPARAERTMRRSDCEFQLVGEPTGEYYEVVMIFGIIDI 731
Query 102 LTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
L + K+ E+A + +Q D IS V+P Y+ RF F+
Sbjct 732 LQDYDISKKLEHAYKSIQY-DPTSISAVDPRLYSRRFRDFI 771
> ath:AT3G09920 PIP5K9; PIP5K9 (PHOSPHATIDYL INOSITOL MONOPHOSPHATE
5 KINASE); 1-phosphatidylinositol-4-phosphate 5-kinase/
ATP binding / phosphatidylinositol phosphate kinase; K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=815
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 34/50 (68%), Gaps = 1/50 (2%)
Query 95 YVGIIDVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFLER 144
Y+GIID+L ++ K+ E+A + + DS IS V+P FY+ RF+ F+++
Sbjct 759 YLGIIDILQEYNMTKKIEHAYKSLH-FDSLSISAVDPTFYSQRFLEFIKK 807
Score = 32.0 bits (71), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 32/65 (49%), Gaps = 7/65 (10%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSFSRVPLPAALTNEPIPSAS 70
LL L D+ FL +MDYSLL+G+++ P S++ ++T + + S +
Sbjct 615 LLRQLEIDSKFLEAQNIMDYSLLLGVHHRA-------PQHLRSQLVRSQSITTDALESVA 667
Query 71 TDTDV 75
D +
Sbjct 668 EDDTI 672
> dre:553629 pip5k1a, MGC110576, zgc:110576; phosphatidylinositol-4-phosphate
5-kinase, type I, alpha (EC:2.7.1.68); K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=559
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 38/142 (26%), Positives = 66/142 (46%), Gaps = 13/142 (9%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIYY-------STSGTAASRPGGSFSRVPLPAALTN 63
L +++D L + +MDYSLLVGI+ +++ ++ G + A+
Sbjct 281 LSRTMQRDCRVLQSFKIMDYSLLVGIHILHRAGEEASTAVPDTQKKGQGQKPLYCTAI-- 338
Query 64 EPIPSASTDTDVPFWRRDLGGLA---SRDGSKLYYVGIIDVLTKWGAFKRTENAIRVVQT 120
E I S P +GG+ S+ L ++GIID+L + K+ E++ + +
Sbjct 339 ESIQGESKSKTSPQPYESMGGIPAFNSKGERLLVFIGIIDILQSYRLVKKLEHSWKALLH 398
Query 121 CDSYGISCVNPAFYASRFISFL 142
D +S P+FYA RF F+
Sbjct 399 -DGDTVSVHRPSFYADRFQKFM 419
> mmu:108083 Pip4k2b, AI848124, PI5P4Kbeta, Pip5k2b, c11; phosphatidylinositol-5-phosphate
4-kinase, type II, beta (EC:2.7.1.149);
K00920 1-phosphatidylinositol-5-phosphate 4-kinase
[EC:2.7.1.149]
Length=416
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 48/177 (27%), Positives = 71/177 (40%), Gaps = 50/177 (28%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIY----------------------- 37
+ +G E LE L++D FL +MDYSLLVGI+
Sbjct 250 LHVGEESKKNFLEKLKRDVEFLAQLKIMDYSLLVGIHDVDRAEQEEMEVEERAEEEECEN 309
Query 38 -------YSTSGTAASRPGG--SFSRVPLPAALTNEPIPSASTDTDVPFWRRDLGGLASR 88
+ GT PG SF R P PS D+ + S
Sbjct 310 DGVGGSLLCSYGTPPDSPGNLLSFPRFFGPGEFD----PSV-----------DVYAMKSH 354
Query 89 DGS---KLYYVGIIDVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
+ + ++Y++ IID+LT + A K+ +A + V+ IS VNP Y+ RF F+
Sbjct 355 ESAPKKEVYFMAIIDILTPYDAKKKAAHAAKTVKHGAGAEISTVNPEQYSKRFNEFM 411
> dre:492549 im:7140933; zgc:158838; K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=584
Score = 45.4 bits (106), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 43/162 (26%), Positives = 71/162 (43%), Gaps = 24/162 (14%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGG---------- 50
I + P+ L + +++D L + +MDYSLLVG++ + ASR
Sbjct 269 IQLEPDNYNALCKTIQRDCLLLQSFKIMDYSLLVGVHNT---DLASRERAGVVEGGGSEG 325
Query 51 ----SFSRVPLPAALTN---EPIPSASTDTDVPFWRRDLGGLASRD--GSK-LYYVGIID 100
R + AL + E I + GG+ +R+ G + L Y+GIID
Sbjct 326 TVTPDHRRPQIQKALYSTAMESIQGEAKGKGTLETEDQWGGIPARNSRGERILVYIGIID 385
Query 101 VLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
+L + K+ E++ + + D +S P FYA RF F+
Sbjct 386 ILQSYRFIKKLEHSWKAL-VHDGDTVSVHRPGFYAERFQRFM 426
> ath:AT3G07960 phosphatidylinositol-4-phosphate 5-kinase family
protein; K00889 1-phosphatidylinositol-4-phosphate 5-kinase
[EC:2.7.1.68]
Length=715
Score = 45.1 bits (105), Expect = 8e-05, Method: Composition-based stats.
Identities = 44/155 (28%), Positives = 68/155 (43%), Gaps = 25/155 (16%)
Query 17 KDANFLVTHGLMDYSLLVGIYY--------STSGTAASRPGGS----FSRVPLPAALTN- 63
+D FL +MDYSLLVG+++ +T + A P G+ SR + L +
Sbjct 559 RDCEFLEQERIMDYSLLVGLHFREAAIKDSATPTSGARTPTGNSETRLSRAEMDRFLLDA 618
Query 64 EPIPSASTDTDVP-----FWRRD------LGGLASRDGSKLYYVGIIDVLTKWGAFKRTE 112
+ S ++P RR +G + Y GIID+L + K+ E
Sbjct 619 SKLASIKLGINMPARVERTARRSDCENQLVGDPTGEFYDVIVYFGIIDILQDYDISKKLE 678
Query 113 NAIRVVQTCDSYGISCVNPAFYASRFISFLERHIV 147
+A + +Q D IS V+P Y+ RF F+ R V
Sbjct 679 HAYKSMQY-DPTSISAVDPKQYSRRFRDFIFRVFV 712
> dre:560360 similar to type II phosphatidylinositolphosphate
kinase-alpha; K00920 1-phosphatidylinositol-5-phosphate 4-kinase
[EC:2.7.1.149]
Length=404
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 46/156 (29%), Positives = 67/156 (42%), Gaps = 14/156 (8%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIY---YSTSGTAASRPGGSFSRVPL 57
I I E L L+KD FL LMDYSLLVGI+ + S
Sbjct 244 IYIDDESKKIFLNKLQKDVEFLALLKLMDYSLLVGIHDVERAEQEEVESEDNEGEEEGES 303
Query 58 PAALTNEP--IPSASTDTDVPFW------RRDLGGLASRDGS---KLYYVGIIDVLTKWG 106
+ P PS + D+ P D+ + S D S ++Y++ +ID+L +
Sbjct 304 DGGIVGTPPDSPSNTLDSTKPLSPGDFDPNIDVYAIKSHDNSPRKEVYFMAVIDILQHYD 363
Query 107 AFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
A K+ +A + V+ IS VNP Y+ RF F+
Sbjct 364 AKKKAAHAAKTVKHGAGAEISTVNPEQYSKRFYDFI 399
> xla:496028 pip4k2c, pip5k2c; phosphatidylinositol-5-phosphate
4-kinase, type II, gamma (EC:2.7.1.149); K00920 1-phosphatidylinositol-5-phosphate
4-kinase [EC:2.7.1.149]
Length=419
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 47/172 (27%), Positives = 75/172 (43%), Gaps = 47/172 (27%)
Query 3 IGPERAARLLELLRKDANFLVTHGLMDYSLLVGIY------------------------Y 38
+ E+ +E L++D +FLV LMDYSLL+GI+ +
Sbjct 258 VDEEQKRNFMEKLKRDVDFLVQLKLMDYSLLLGIHEVFRAEQEEEEELEEDNAENECSPH 317
Query 39 STSGTAASRPGGSF----SRVPL-PAALTNEPIPSASTDTDVPFWRRDLGGLASRDGS-- 91
G+ + P G S PL P EPI D+ + S D +
Sbjct 318 VNVGSYGTSPEGIAGYLNSHKPLGPGEF--EPII-------------DVYAIKSSDNAPQ 362
Query 92 -KLYYVGIIDVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
++Y++G+ID+LT + A K+ +A + V+ IS V+P Y RF+ F+
Sbjct 363 KEVYFMGLIDILTHYDAKKKAAHAAKTVKHGAGAEISTVHPDQYGKRFLEFV 414
> ath:AT1G01460 PIPK11; PIPK11; 1-phosphatidylinositol-4-phosphate
5-kinase/ phosphatidylinositol phosphate kinase; K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=427
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 37/66 (56%), Gaps = 6/66 (9%)
Query 84 GLASRDGSK-----LYYVGIIDVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRF 138
G SR+G + + Y+GIID+ +G KR E+ + +Q S IS V+P Y+SRF
Sbjct 353 GKQSREGGEEWYDVILYLGIIDIFQDYGVRKRLEHCYKSIQHS-SKTISAVHPKMYSSRF 411
Query 139 ISFLER 144
F+ +
Sbjct 412 QDFVSQ 417
> ath:AT2G41210 PIP5K5; PIP5K5 (PHOSPHATIDYLINOSITOL- 4-PHOSPHATE
5-KINASE 5); 1-phosphatidylinositol-4-phosphate 5-kinase;
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=772
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 43/153 (28%), Positives = 66/153 (43%), Gaps = 28/153 (18%)
Query 17 KDANFLVTHGLMDYSLLVGIYYSTSGTAASR-PGGSFSRVPLPAALTNEPIPSASTD--- 72
KD FL +MDYSLLVGI++ + A P G+ + + + + A D
Sbjct 613 KDCEFLEQERIMDYSLLVGIHFREASVAGELIPSGARTPIGESEEESGPRLSRAEVDELL 672
Query 73 TDVPFWRR-----DLGGLASR----DGSKLYYVG--------------IIDVLTKWGAFK 109
+D W ++ A R + S+L VG IID+L + K
Sbjct 673 SDPSRWASIRLGTNMPARAERTMRKNDSELQLVGEPTGEFYEVVMIFGIIDILQDYDISK 732
Query 110 RTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
+ E+A + +Q D IS V+P Y+ RF F+
Sbjct 733 KLEHAYKSIQY-DPSSISAVDPRQYSRRFRDFI 764
> hsa:23396 PIP5K1C, KIAA0589, LCCS3, PIP5K-GAMMA, PIP5Kgamma,
PIPKIg_v4; phosphatidylinositol-4-phosphate 5-kinase, type
I, gamma (EC:2.7.1.68); K00889 1-phosphatidylinositol-4-phosphate
5-kinase [EC:2.7.1.68]
Length=640
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 39/154 (25%), Positives = 65/154 (42%), Gaps = 26/154 (16%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIYY------------STSGTAASRPGGSFSRVPLP 58
L++ L++D L + +MDYSLL+G++ + S + RP G +
Sbjct 298 LVKTLQRDCLVLESFKIMDYSLLLGVHNIDQHERERQAQGAQSTSDEKRPVGQKALYSTA 357
Query 59 ------AALTNEPIPSASTDTDVPFWRRDLGGLASRDGSKLYYVGIIDVLTKWGAFKRTE 112
A E I S T +P + R L ++GIID+L + K+ E
Sbjct 358 MESIQGGAARGEAIESDDTMGGIP-------AVNGRGERLLLHIGIIDILQSYRFIKKLE 410
Query 113 NAIRVVQTCDSYGISCVNPAFYASRFISFLERHI 146
+ + + D +S P+FYA RF F+ +
Sbjct 411 HTWKAL-VHDGDTVSVHRPSFYAERFFKFMSNTV 443
> mmu:18720 Pip5k1a, Pip5k1b, Pipk5a, Pipk5b; phosphatidylinositol-4-phosphate
5-kinase, type 1 alpha (EC:2.7.1.68); K00889
1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=546
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 38/143 (26%), Positives = 64/143 (44%), Gaps = 12/143 (8%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGI----YYSTSGTAASRPGGSFSRVPLPA----ALT 62
L + L++D L + +MDYSLL+ I + T+ + +R P P +
Sbjct 289 LCKTLQRDCLVLQSFKIMDYSLLMSIHNMDHAQREPTSNDTQYSADTRRPAPQKALYSTA 348
Query 63 NEPIPSASTDTDVPFWRRDLGGLASRDGSK---LYYVGIIDVLTKWGAFKRTENAIRVVQ 119
E I + +GG+ +R+ L Y+GIID+L + K+ E++ + +
Sbjct 349 MESIQGEARRGGTVETEDHMGGIPARNNKGERLLLYIGIIDILQSYRFVKKLEHSWKAL- 407
Query 120 TCDSYGISCVNPAFYASRFISFL 142
D +S P FYA RF F+
Sbjct 408 VHDGDTVSVHRPGFYAERFQRFM 430
> hsa:138429 PIP5KL1, MGC46424, PIPKH, bA203J24.5; phosphatidylinositol-4-phosphate
5-kinase-like 1 (EC:2.7.1.68); K13712
phosphatidylinositol-4-phosphate 5-kinase-like protein 1 [EC:2.7.1.68]
Length=394
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 41/150 (27%), Positives = 65/150 (43%), Gaps = 14/150 (9%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSFS-RVPLPA 59
I++GP+R+ L + D FL ++DYSLL+ + PG S R
Sbjct 252 INLGPQRSW-FLRQMELDTTFLRELNVLDYSLLIA--FQRLHEDERGPGSSLIFRTARSV 308
Query 60 ALTNEPIPSASTDT----DVPFWRRDLGGLASRDGSKLYYVGIIDVLTKWGAFKRTENAI 115
P S + + D P L G R Y++G++D+ T +G KR E+
Sbjct 309 QGAQSPEESRAQNRRLLPDAPNALHILDGPEQR-----YFLGVVDLATVYGLRKRLEHLW 363
Query 116 RVVQTCDSYGISCVNPAFYASRFISFLERH 145
+ ++ S V+PA YA R ++E H
Sbjct 364 KTLR-YPGRTFSTVSPARYARRLCQWVEAH 392
> mmu:227733 Pip5kl1, BC028795; phosphatidylinositol-4-phosphate
5-kinase-like 1 (EC:2.7.1.68); K13712 phosphatidylinositol-4-phosphate
5-kinase-like protein 1 [EC:2.7.1.68]
Length=395
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 44/156 (28%), Positives = 69/156 (44%), Gaps = 27/156 (17%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLV----------GIYYSTSGTAASRPGG 50
+ +G +R+ L + D FL ++DYSLLV GI++S T S G
Sbjct 254 MRLGAQRSW-FLRQMELDTAFLREVNVLDYSLLVAIQFLHEDEKGIHHSVFSTFKSIQGV 312
Query 51 SFSRVPLPAALTNEP-IPSASTDTDVPFWRRDLGGLASRDGSKLYYVGIIDVLTKWGAFK 109
S S+ P +P+A D P R Y++G++D+ T +G K
Sbjct 313 SKSKGTGDQNCRMLPDLPNALHILDGPDQR--------------YFLGLVDMTTVYGFRK 358
Query 110 RTENAIRVVQTCDSYGISCVNPAFYASRFISFLERH 145
R E+ ++V+ +S V+PA YA R + E H
Sbjct 359 RLEHVWKMVR-YPGQSVSTVSPAHYARRLCRWAEVH 393
> ath:AT4G01190 ATPIPK10 (Arabidopsis phosphatidylinositol phosphate
kinase 10); 1-phosphatidylinositol-4-phosphate 5-kinase;
K00889 1-phosphatidylinositol-4-phosphate 5-kinase [EC:2.7.1.68]
Length=401
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/48 (39%), Positives = 29/48 (60%), Gaps = 1/48 (2%)
Query 95 YVGIIDVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
Y+GI+D +G KR E+ + +Q +S IS V+P Y+SRF F+
Sbjct 340 YIGIVDTFQDYGMKKRIEHCYKSIQY-NSNSISTVHPKIYSSRFQDFV 386
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 3 IGPERAARLLELLRKDANFLVTHGLMDYSLLVGI 36
+ P R+++ + D L G+MDYSLLVG+
Sbjct 184 VDPLARQRIIKQTKLDCELLEEEGIMDYSLLVGL 217
> ath:AT1G60890 phosphatidylinositol-4-phosphate 5-kinase family
protein; K00889 1-phosphatidylinositol-4-phosphate 5-kinase
[EC:2.7.1.68]
Length=769
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 29/48 (60%), Gaps = 1/48 (2%)
Query 95 YVGIIDVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFL 142
Y+GIID+L ++ K+ E+ + ++ D IS + P Y+ RFI FL
Sbjct 715 YMGIIDILQEYNMKKKVEHTCKSMK-YDPMTISAIEPTLYSKRFIDFL 761
> tgo:TGME49_045730 phosphatidylinositol-4-phosphate 5-Kinase,
putative (EC:2.7.1.68)
Length=2153
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 35/62 (56%), Gaps = 3/62 (4%)
Query 89 DGSKLYYVGIIDVLT--KWGAFKRTE-NAIRVVQTCDSYGISCVNPAFYASRFISFLERH 145
D +++YY+GIID LT W + T ++ C+ IS V PA+Y+ R +FL H
Sbjct 1904 DMTEIYYLGIIDFLTPYDWRRYGHTVYKRLKSSAKCEFGEISPVPPAYYSRRQQAFLRDH 1963
Query 146 IV 147
+V
Sbjct 1964 VV 1965
Score = 32.0 bits (71), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 1 IDIGPERAARLLELLRKDANFLVTHGLMDYSLLVGIY 37
+++ E A +L R D FL T + DYSLLVG++
Sbjct 1444 LNMRDEEAQTILRAHRLDCEFLETISVFDYSLLVGVH 1480
> ath:AT1G10900 phosphatidylinositol-4-phosphate 5-kinase family
protein; K00889 1-phosphatidylinositol-4-phosphate 5-kinase
[EC:2.7.1.68]
Length=754
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 31/50 (62%), Gaps = 1/50 (2%)
Query 95 YVGIIDVLTKWGAFKRTENAIRVVQTCDSYGISCVNPAFYASRFISFLER 144
Y+GIID+L ++ K+ E+ + +Q D IS P+ Y+ RF++FL +
Sbjct 700 YMGIIDILQEYNTKKKVEHTCKSLQY-DPMTISVTEPSTYSKRFVNFLHK 748
> dre:100149843 phosphatidylinositol-4-phosphate 5-kinase, type
I, alpha-like; K13712 phosphatidylinositol-4-phosphate 5-kinase-like
protein 1 [EC:2.7.1.68]
Length=388
Score = 35.0 bits (79), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 37/140 (26%), Positives = 59/140 (42%), Gaps = 8/140 (5%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGIYYSTSGTAASRPGGSFSRVPLPAALTNEPIPSAS 70
LL + D FL T ++DYSLL+ + SF+ + + SA
Sbjct 250 LLRQVEIDTAFLQTLNVLDYSLLLA--HQPLHQDELSQSLSFATLIMRTKKNVAGAESAD 307
Query 71 TDTDVPFWRRDLGGLASR----DG-SKLYYVGIIDVLTKWGAFKRTENAIRVVQTCDSYG 125
RR L + DG + Y++GIID+ T +G KR E+ + ++ S
Sbjct 308 LQAFQSQNRRLLPNFKNPLHVIDGPEQRYFIGIIDIFTVYGFRKRLEHLWKRLRH-PSQT 366
Query 126 ISCVNPAFYASRFISFLERH 145
S V+P Y R +++ H
Sbjct 367 FSTVSPPAYCHRLCHWVQDH 386
> dre:561989 similar to galanin receptor type 1; K04230 galanin
receptor 1
Length=350
Score = 32.3 bits (72), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query 98 IIDVLTKWGAFKRTEN--AIRVVQTCDSYGISCVNPAFYA 135
II + ++G F + A R++ C +YG SCVNP YA
Sbjct 266 IIAMWVEFGQFPLNDASFAFRIISHCLAYGNSCVNPILYA 305
> pfa:PF14_0123 conserved Plasmodium protein, unknown function
Length=3218
Score = 32.0 bits (71), Expect = 0.63, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 11 LLELLRKDANFLVTHGLMDYSLLVGI 36
L++ L +D NFL + +MDYSLL+ I
Sbjct 2751 LMDSLNEDTNFLCSQDIMDYSLLIHI 2776
Lambda K H
0.322 0.139 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40