bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2116_orf1
Length=181
Score E
Sequences producing significant alignments: (Bits) Value
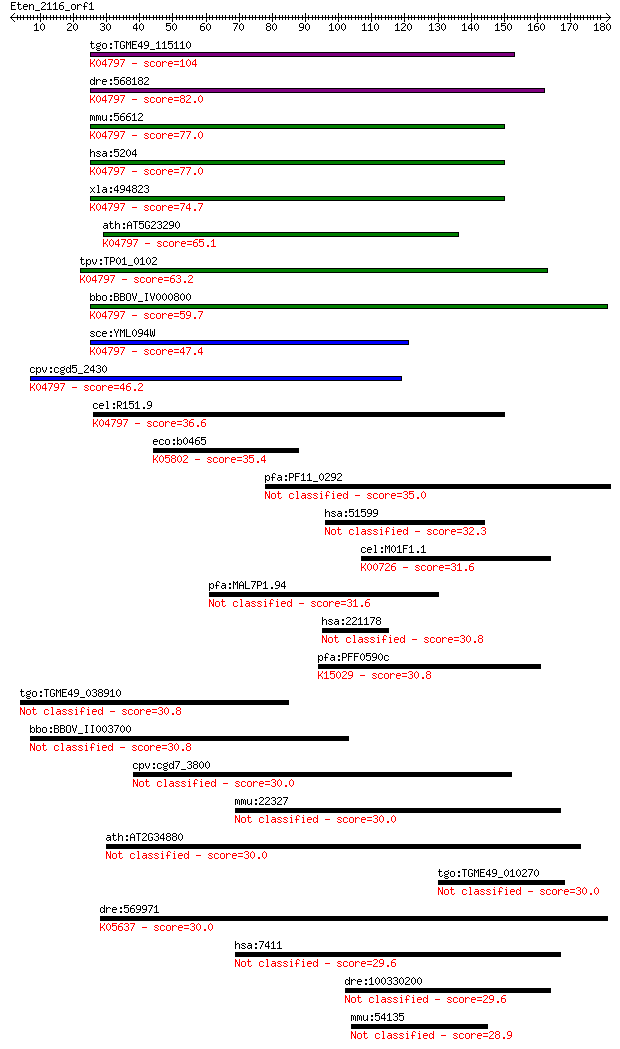
tgo:TGME49_115110 prefoldin subunit 5, putative ; K04797 prefo... 104 1e-22
dre:568182 pfdn5, wu:fa09c11, wu:fb38c05, zgc:100894; prefoldi... 82.0 1e-15
mmu:56612 Pfdn5, 1190001O17Rik, 1700010A06Rik, D15Ertd697e, Ei... 77.0 3e-14
hsa:5204 PFDN5, MGC5329, MGC71907, MM-1, MM1, PFD5; prefoldin ... 77.0 3e-14
xla:494823 pfdn5; prefoldin subunit 5; K04797 prefoldin alpha ... 74.7 1e-13
ath:AT5G23290 PDF5; PDF5 (PREFOLDIN 5); unfolded protein bindi... 65.1 1e-10
tpv:TP01_0102 prefoldin subunit 5; K04797 prefoldin alpha subunit 63.2 5e-10
bbo:BBOV_IV000800 21.m02738; prefoldin subunit 5 (c-myc bindin... 59.7 5e-09
sce:YML094W GIM5, PFD5; Gim5p; K04797 prefoldin alpha subunit 47.4 3e-05
cpv:cgd5_2430 hypothetical protein ; K04797 prefoldin alpha su... 46.2 7e-05
cel:R151.9 pfd-5; PreFolDin (molecular chaperone) family membe... 36.6 0.053
eco:b0465 kefA, aefA, ECK0459, JW0454, mscK, pnuH; fused conse... 35.4 0.11
pfa:PF11_0292 conserved Plasmodium protein 35.0 0.13
hsa:51599 LSR, ILDR3, LISCH7, MGC10659, MGC48312, MGC48503; li... 32.3 0.86
cel:M01F1.1 gly-14; GLYcosylation related family member (gly-1... 31.6 1.4
pfa:MAL7P1.94 prefoldin subunit 3, putative 31.6 1.6
hsa:221178 SPATA13, ARHGEF29, ASEF2, FLJ31208, FLJ35435, MGC12... 30.8 2.4
pfa:PFF0590c homologue of human HSPC025; K15029 translation in... 30.8 2.7
tgo:TGME49_038910 hypothetical protein 30.8 2.7
bbo:BBOV_II003700 18.m06311; LCCL domain-containing protein CCP2 30.8 2.9
cpv:cgd7_3800 hypothetical protein 30.0 4.0
mmu:22327 Vbp1; von Hippel-Lindau binding protein 1 30.0
ath:AT2G34880 MEE27; MEE27 (maternal effect embryo arrest 27);... 30.0 4.4
tgo:TGME49_010270 hypothetical protein 30.0 4.4
dre:569971 lama1; laminin, alpha 1; K05637 laminin, alpha 1/2 30.0
hsa:7411 VBP1, PFD3, PFDN3, VBP-1; von Hippel-Lindau binding p... 29.6 5.4
dre:100330200 dock2; dedicator of cytokinesis 2 29.6
mmu:54135 Lsr, ILDR3, Lisch7; lipolysis stimulated lipoprotein... 28.9 9.4
> tgo:TGME49_115110 prefoldin subunit 5, putative ; K04797 prefoldin
alpha subunit
Length=178
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 55/128 (42%), Positives = 87/128 (67%), Gaps = 0/128 (0%)
Query 25 NLGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEA 84
+L SL+L Q + + +AEV+NLS HL L+ A+ R+ EAREA+ + + + +E +
Sbjct 18 SLASLSLQQLVGVKEQLDAEVQNLSAHLRTLRMASGRLQEAREALSKFASPAEATEEEDQ 77
Query 85 PEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEK 144
EVLVPLT ++YVKGRL K+LVD+G G++I+K +D KK R ++ ++EQ A++EK
Sbjct 78 AEVLVPLTSSVYVKGRLVTRKKLLVDVGTGFLIEKNCEDAKKGLDRNVSMVNEQVAKVEK 137
Query 145 IISDKVKQ 152
I+ +K +Q
Sbjct 138 ILPEKQRQ 145
> dre:568182 pfdn5, wu:fa09c11, wu:fb38c05, zgc:100894; prefoldin
5; K04797 prefoldin alpha subunit
Length=153
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 46/137 (33%), Positives = 79/137 (57%), Gaps = 6/137 (4%)
Query 25 NLGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEA 84
NL L+L Q L + + E E LS+ ++QLK ++ EA++++ L+ K E
Sbjct 4 NLTELSLPQLEGLKTQLDQETEFLSSSIAQLKVVQTKYVEAKDSLNVLN------KSNEG 57
Query 85 PEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEK 144
E+LVPLT ++YV G+L D VLVD+G GY ++K +D K+ R + F+++Q +++
Sbjct 58 KELLVPLTSSMYVPGKLHDVDHVLVDVGTGYFVEKNVEDGKEFFKRKIDFLTKQIEKIQP 117
Query 145 IISDKVKQLQVLVATLN 161
+ +K Q +V +N
Sbjct 118 ALQEKYAMKQAVVEVMN 134
> mmu:56612 Pfdn5, 1190001O17Rik, 1700010A06Rik, D15Ertd697e,
Eig1, MM-1; prefoldin 5; K04797 prefoldin alpha subunit
Length=154
Score = 77.0 bits (188), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 42/125 (33%), Positives = 72/125 (57%), Gaps = 6/125 (4%)
Query 25 NLGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEA 84
N+ L L Q L + + EVE LS ++QLK ++ EA++ + L+ K E
Sbjct 6 NITELNLPQLEMLKNQLDQEVEFLSTSIAQLKVVQTKYVEAKDCLNVLN------KSNEG 59
Query 85 PEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEK 144
E+LVPLT ++YV G+L + VL+D+G GY ++KT +D K R + F+++Q +++
Sbjct 60 KELLVPLTSSMYVPGKLHDVEHVLIDVGTGYYVEKTAEDAKDFFKRKIDFLTKQMEKIQP 119
Query 145 IISDK 149
+ +K
Sbjct 120 ALQEK 124
> hsa:5204 PFDN5, MGC5329, MGC71907, MM-1, MM1, PFD5; prefoldin
subunit 5; K04797 prefoldin alpha subunit
Length=154
Score = 77.0 bits (188), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 42/125 (33%), Positives = 72/125 (57%), Gaps = 6/125 (4%)
Query 25 NLGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEA 84
N+ L L Q L + + EVE LS ++QLK ++ EA++ + L+ K E
Sbjct 6 NITELNLPQLEMLKNQLDQEVEFLSTSIAQLKVVQTKYVEAKDCLNVLN------KSNEG 59
Query 85 PEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEK 144
E+LVPLT ++YV G+L + VL+D+G GY ++KT +D K R + F+++Q +++
Sbjct 60 KELLVPLTSSMYVPGKLHDVEHVLIDVGTGYYVEKTAEDAKDFFKRKIDFLTKQMEKIQP 119
Query 145 IISDK 149
+ +K
Sbjct 120 ALQEK 124
> xla:494823 pfdn5; prefoldin subunit 5; K04797 prefoldin alpha
subunit
Length=152
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 40/125 (32%), Positives = 73/125 (58%), Gaps = 6/125 (4%)
Query 25 NLGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEA 84
N+ L+L Q L + + EVE LS+ ++QLK ++ EA+E + L+ K E
Sbjct 6 NITDLSLPQLEGLKSQLDQEVEFLSSSIAQLKVVQTKYVEAKECLSVLN------KSNEG 59
Query 85 PEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEK 144
++LVPLT ++YV G L+ +++D+G GY ++KT +D K R + F+++Q +++
Sbjct 60 KQLLVPLTSSMYVPGTLNDVSTIMIDVGTGYYVEKTVEDAKDFFKRKVEFLTKQIEKIQP 119
Query 145 IISDK 149
+ +K
Sbjct 120 ALQEK 124
> ath:AT5G23290 PDF5; PDF5 (PREFOLDIN 5); unfolded protein binding;
K04797 prefoldin alpha subunit
Length=151
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 42/107 (39%), Positives = 59/107 (55%), Gaps = 6/107 (5%)
Query 29 LTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEAPEVL 88
+ +DQ +L +A+ EV L + L+ ++ A R+ A A+ L Q +K +L
Sbjct 13 MGIDQLKALKEQADLEVNLLQDSLNNIRTATVRLDAAAAALNDLSLRPQGKK------ML 66
Query 89 VPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFM 135
VPLT +LYV G LD DKVLVDIG GY I+KT D K R + +
Sbjct 67 VPLTASLYVPGTLDEADKVLVDIGTGYFIEKTMDDGKDYCQRKINLL 113
> tpv:TP01_0102 prefoldin subunit 5; K04797 prefoldin alpha subunit
Length=164
Score = 63.2 bits (152), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 74/141 (52%), Gaps = 8/141 (5%)
Query 22 DINNLGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKE 81
D L + ++ + + L+ + E EV L + ++ L A R E+++A+ +L E
Sbjct 16 DARRLENFSIQELNMLILKLEEEVNELQSLVNALTIAMERFQESKKALTEL--------E 67
Query 82 GEAPEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQAR 141
+ ++ VPLT +YV G L DKVLV +G GY ++ + + R + + EQ +
Sbjct 68 KKNKQIQVPLTSLVYVPGELTNPDKVLVSVGTGYYVEMDLKKAEDYYERRIRTVEEQMCK 127
Query 142 LEKIISDKVKQLQVLVATLNR 162
L+ I+S K KQ+ + + L++
Sbjct 128 LQAILSGKAKQINLSYSYLDQ 148
> bbo:BBOV_IV000800 21.m02738; prefoldin subunit 5 (c-myc binding
protein); K04797 prefoldin alpha subunit
Length=167
Score = 59.7 bits (143), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 40/156 (25%), Positives = 76/156 (48%), Gaps = 8/156 (5%)
Query 25 NLGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEA 84
N+ +L++ + + ++ R E E+ + N ++ L A R+ E++ + + ++
Sbjct 19 NVHNLSIQELNMIILRLEEEINQMQNLVNVLTVALERLHESKACL--------KDFSSQS 70
Query 85 PEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEK 144
E+ VPLT +YV G++ KVLV +G GY I+ T + R + + EQ +L
Sbjct 71 CEIQVPLTSLVYVPGKIANPGKVLVSVGTGYFIEMTLDKAGEFYERKIEVVEEQLRKLHS 130
Query 145 IISDKVKQLQVLVATLNRSRDEAPSGASVARRQPAS 180
I S K KQ+ + L+ + + + A Q A+
Sbjct 131 ICSAKNKQITQTYSVLDHKLSQLRNAQAAATSQVAT 166
> sce:YML094W GIM5, PFD5; Gim5p; K04797 prefoldin alpha subunit
Length=163
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 52/96 (54%), Gaps = 5/96 (5%)
Query 25 NLGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEA 84
+L L +Q +++ ++ + E+++ + L L A + +E + D Q E
Sbjct 7 DLTKLNPEQLNAVKQQFDQELQHFTQSLQALTMAKGKFTECID-----DIKTVSQAGNEG 61
Query 85 PEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKT 120
++LVP + +LY+ G++ + K +VDIG GY ++K+
Sbjct 62 QKLLVPASASLYIPGKIVDNKKFMVDIGTGYYVEKS 97
> cpv:cgd5_2430 hypothetical protein ; K04797 prefoldin alpha
subunit
Length=163
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 34/113 (30%), Positives = 56/113 (49%), Gaps = 8/113 (7%)
Query 7 LCEAMAAPTAGTVKQDINNLGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAR 66
+ A AP K + L SL + L + + E+ LS + QL +R + +R
Sbjct 1 IMTASNAPNNSGNKVETIQLSSLPPAKLFQLRDQTQDEMNELSIRIQQLNVVLNRFNGSR 60
Query 67 EAVIQLDAYRQRQKEGEAPEVLVPLTGALYVKGRLDCD-DKVLVDIGAGYMIQ 118
EA+ QL + E + +L P++ ++YV + CD + VLVDIG GY ++
Sbjct 61 EALEQL------KPENKDNTILAPISQSIYVDATI-CDVENVLVDIGTGYHVE 106
> cel:R151.9 pfd-5; PreFolDin (molecular chaperone) family member
(pfd-5); K04797 prefoldin alpha subunit
Length=152
Score = 36.6 bits (83), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 32/124 (25%), Positives = 56/124 (45%), Gaps = 6/124 (4%)
Query 26 LGSLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEAP 85
L L+L Q L + E E+ + LK +R +++ LD + A
Sbjct 10 LSELSLQQLGELQKNCEQELNFFQESFNALKGLLTR---NEKSISALDDVKIATAGHTA- 65
Query 86 EVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEKI 145
L+PL+ +LY++ L K LV+IG GY ++ + K R +++Q +E I
Sbjct 66 --LIPLSESLYIRAELSDPSKHLVEIGTGYFVELDREKAKAIFDRKKEHITKQVETVEGI 123
Query 146 ISDK 149
+ +K
Sbjct 124 LKEK 127
> eco:b0465 kefA, aefA, ECK0459, JW0454, mscK, pnuH; fused conserved
protein; K05802 potassium efflux system protein KefA
Length=1120
Score = 35.4 bits (80), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 28/44 (63%), Gaps = 0/44 (0%)
Query 44 EVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEAPEV 87
E+EN++N ++ L+ V++ R+A+ Q DA+ + +EG EV
Sbjct 361 ELENMTNRIADLRLEQFEVNQQRDALFQSDAFVNKLEEGHTNEV 404
> pfa:PF11_0292 conserved Plasmodium protein
Length=248
Score = 35.0 bits (79), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 23/111 (20%), Positives = 54/111 (48%), Gaps = 14/111 (12%)
Query 78 RQKEGEAPEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTF--- 134
++ E ++ +V +PLT +Y+ G++ D LV +G Y ++ +++ +++ +
Sbjct 145 KEDEKKSFDVHIPLTSLVYIPGKIVNTDNFLVRMGTNYYVE-------RNSTQVIEYYNN 197
Query 135 ----MSEQQARLEKIISDKVKQLQVLVATLNRSRDEAPSGASVARRQPASA 181
++EQ +L+ I +K ++ + + R E A +QP S+
Sbjct 198 KIKKINEQITKLKITIIEKKNEIDLCKNYIQIKRREQQMNALNMPQQPTSS 248
> hsa:51599 LSR, ILDR3, LISCH7, MGC10659, MGC48312, MGC48503;
lipolysis stimulated lipoprotein receptor
Length=630
Score = 32.3 bits (72), Expect = 0.86, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 27/48 (56%), Gaps = 3/48 (6%)
Query 96 YVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLE 143
YV D D V ++ +GY IQ + QD D+MR+L +M ++ A +
Sbjct 353 YVPLLRDTDSSVASEVRSGYRIQASQQD---DSMRVLYYMEKELANFD 397
> cel:M01F1.1 gly-14; GLYcosylation related family member (gly-14);
K00726 alpha-1,3-mannosyl-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
[EC:2.4.1.101]
Length=437
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Query 107 VLVDIGAGYMIQKTYQDEK--KDAMRILTFMSEQQARLEKIISDKVKQLQVLVATLNRS 163
VL+ + IQ + ++ K+ +R+L M E +RLEK++ + + ++ L T+NR+
Sbjct 12 VLIYFSWNFYIQNGFTNKAIVKEELRLLKEMDEAGSRLEKLLKVEKEHMERLSETMNRA 70
> pfa:MAL7P1.94 prefoldin subunit 3, putative
Length=192
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 41/71 (57%), Gaps = 3/71 (4%)
Query 61 RVSEAREA--VIQLDAYRQRQKEGEAPEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQ 118
++ E ++A V+QL R++ KE + E P+ +LY +G + DK+L+ +GA M++
Sbjct 65 KIPELKDALRVVQLLEKRKKIKEKKNLETFFPVEESLYARGIVQKTDKILLWLGANVMVE 124
Query 119 KTYQDEKKDAM 129
+ DE D +
Sbjct 125 FPF-DEATDLL 134
> hsa:221178 SPATA13, ARHGEF29, ASEF2, FLJ31208, FLJ35435, MGC129988,
MGC129989; spermatogenesis associated 13
Length=1277
Score = 30.8 bits (68), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 13/20 (65%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 95 LYVKGRLDCDDKVLVDIGAG 114
LY KGRLD D+ LVD+G G
Sbjct 1121 LYYKGRLDMDEMELVDLGDG 1140
> pfa:PFF0590c homologue of human HSPC025; K15029 translation
initiation factor 3 subunit L
Length=656
Score = 30.8 bits (68), Expect = 2.7, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 34/67 (50%), Gaps = 5/67 (7%)
Query 94 ALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEKIISDKVKQL 153
ALY K L C + +I YM+ K Y D K +IL ++S+Q+ +S++ K
Sbjct 265 ALYWKVTL-CHLSIFYNIAFSYMMLKRYNDSIKVLSQILIYLSKQKIH----VSNQQKYQ 319
Query 154 QVLVATL 160
Q L+ L
Sbjct 320 QNLIHKL 326
> tgo:TGME49_038910 hypothetical protein
Length=782
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 42/85 (49%), Gaps = 4/85 (4%)
Query 4 SFMLCEAMAAPTAGT-VKQDINNLGSLTLDQCSSLVRRA---EAEVENLSNHLSQLKFAA 59
SF E + P A + ++ +G +TLDQ + ++ + EA +E L N + KFA+
Sbjct 27 SFGASELLKTPAASLEGAEGLHVIGFVTLDQLVASLKDSSLREAFLEALPNQWALPKFAS 86
Query 60 SRVSEAREAVIQLDAYRQRQKEGEA 84
S SE + + R+R G+A
Sbjct 87 STTSELNQLQAWTKSIRERFPLGDA 111
> bbo:BBOV_II003700 18.m06311; LCCL domain-containing protein
CCP2
Length=1597
Score = 30.8 bits (68), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 47/100 (47%), Gaps = 8/100 (8%)
Query 7 LCEAMAAPTAGTVKQDINNLGSLTLDQCSSLVRRAEAEVE-NLSNHLSQLKFAASRVSEA 65
LC + A +AG +N + + +L + AE+ V+ NL ++ + F A + +
Sbjct 368 LCVSQAGSSAGLANVALNKIATSSLPMKNDKSHNAESAVDGNLQSYWASESFTADSIPDK 427
Query 66 REAVIQL-DAYRQR--QKEGEAPEVLVPLTGALYVKGRLD 102
E ++ L + Y+ R + E EAP LT +Y K D
Sbjct 428 VEFIVNLQEQYKIRKLEIEWEAPA----LTYNIYTKLETD 463
> cpv:cgd7_3800 hypothetical protein
Length=2901
Score = 30.0 bits (66), Expect = 4.0, Method: Composition-based stats.
Identities = 24/114 (21%), Positives = 49/114 (42%), Gaps = 1/114 (0%)
Query 38 VRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEAPEVLVPLTGALYV 97
+R+ E ++++ + + + V E + + + + +KE +AP+V + +T L V
Sbjct 1756 IRQKETFLDDIPETIPSIWPSIGTVVELEKGIDE-EGAELNKKEEKAPKVNILITSELEV 1814
Query 98 KGRLDCDDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEKIISDKVK 151
K R +CD + + Q R+L E ++ KI S +K
Sbjct 1815 KSRFNCDGLFVWQVNRAVYWAGMLQRYINKGPRLLKDRKELLKKISKIWSINIK 1868
> mmu:22327 Vbp1; von Hippel-Lindau binding protein 1
Length=196
Score = 30.0 bits (66), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 43/99 (43%), Gaps = 15/99 (15%)
Query 69 VIQLDAYRQRQKEG-EAPEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKD 127
+++ Y Q++KE + E L LY K + DKV + +GA M++
Sbjct 83 TLEILKYMQKKKESTNSMETRFLLADNLYCKASVPPTDKVCLWLGANVMLEYD------- 135
Query 128 AMRILTFMSEQQARLEKIISDKVKQLQVLVATLNRSRDE 166
+ E QA LEK +S K L L L+ RD+
Sbjct 136 -------IDEAQALLEKNLSTATKNLDSLEEDLDFLRDQ 167
> ath:AT2G34880 MEE27; MEE27 (maternal effect embryo arrest 27);
transcription factor
Length=806
Score = 30.0 bits (66), Expect = 4.4, Method: Composition-based stats.
Identities = 42/168 (25%), Positives = 67/168 (39%), Gaps = 41/168 (24%)
Query 30 TLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEAPEVLV 89
T+D+ SSLVR E E ++L LS++ S + + I + KE + E
Sbjct 559 TIDELSSLVRALEGESDDLKAWLSKVMEGCSETQKGESSGIIV-------KEKQVQEECF 611
Query 90 PLTGALYVKGRLDCDDKVLVDIGA--------GYMI------------QKTYQDEKK--- 126
L G + C+D ++D+ A G+++ K ++ K
Sbjct 612 DLNGECNKSSEI-CEDASIMDLAAYHVEPINLGFLVVGKLWCNKHAIFPKGFKSRVKFYN 670
Query 127 --DAMRILTFMSEQQARLEKIISDKVKQLQVLVATLNRSRDEAPSGAS 172
D MRI ++SE I D + TL S+DE+ S AS
Sbjct 671 VQDPMRISYYVSE--------IVDAGLLGPLFKVTLEESQDESFSYAS 710
> tgo:TGME49_010270 hypothetical protein
Length=1897
Score = 30.0 bits (66), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 130 RILTFMSEQQARLEKIISDKVKQLQVLVATLNRSRDEA 167
R + F+S Q+A +E ++V+ L+ VATL R R EA
Sbjct 538 REIEFVSLQKAEVESAAEERVRFLETQVATLQRERAEA 575
> dre:569971 lama1; laminin, alpha 1; K05637 laminin, alpha 1/2
Length=3075
Score = 30.0 bits (66), Expect = 5.0, Method: Composition-based stats.
Identities = 40/168 (23%), Positives = 71/168 (42%), Gaps = 16/168 (9%)
Query 28 SLTLDQCSSLVRRAEAEVENLSNHLSQLKFAASRVSEAREAVIQLDAYRQRQKEGEAPEV 87
S LD+ + L + ++N+S S+L A+ + + + + + EG E+
Sbjct 1953 SEILDRAALLKNHTDGLMQNVSAVTSRLSLVAANIQNFTQMLPEALGILRNLPEGSRAEI 2012
Query 88 L----------VPLTGAL--YVKGRLDCDDKVLVDIGAGYMIQKTYQ---DEKKDAMRIL 132
L L AL RL D+ A Q+T Q D + A
Sbjct 2013 LQVKQQTEAANASLQEALERLRDYRLKLDESSSALGSADNSTQRTNQLLRDSQDTANAAG 2072
Query 133 TFMSEQQARLEKIISDKVKQLQVLVATLNRSRDEAPSGASVARRQPAS 180
+ +SE + ++++ +++K LQ L TL+R+ + S AR+Q AS
Sbjct 2073 SRLSEADVKADRLL-ERLKPLQTLGETLSRNLSDIRELISQARKQAAS 2119
> hsa:7411 VBP1, PFD3, PFDN3, VBP-1; von Hippel-Lindau binding
protein 1
Length=197
Score = 29.6 bits (65), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 43/99 (43%), Gaps = 15/99 (15%)
Query 69 VIQLDAYRQRQKEG-EAPEVLVPLTGALYVKGRLDCDDKVLVDIGAGYMIQKTYQDEKKD 127
+++ Y Q++KE + E L LY K + DKV + +GA M++
Sbjct 84 TLEILKYMQKKKESTNSMETRFLLADNLYCKASVPPTDKVCLWLGANVMLEYD------- 136
Query 128 AMRILTFMSEQQARLEKIISDKVKQLQVLVATLNRSRDE 166
+ E QA LEK +S K L L L+ RD+
Sbjct 137 -------IDEAQALLEKNLSTATKNLDSLEEDLDFLRDQ 168
> dre:100330200 dock2; dedicator of cytokinesis 2
Length=594
Score = 29.6 bits (65), Expect = 6.2, Method: Composition-based stats.
Identities = 18/66 (27%), Positives = 37/66 (56%), Gaps = 6/66 (9%)
Query 102 DCDD----KVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEKIISDKVKQLQVLV 157
DC D +L ++ G + + QDEKK+++ +L + E +R + + D + +Q +V
Sbjct 514 DCRDVLLPMMLRELAGGLALMEGLQDEKKNSIELLNNILEVLSRSD--VGDTFQHIQDIV 571
Query 158 ATLNRS 163
++L R+
Sbjct 572 SSLLRT 577
> mmu:54135 Lsr, ILDR3, Lisch7; lipolysis stimulated lipoprotein
receptor
Length=575
Score = 28.9 bits (63), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 23/41 (56%), Gaps = 3/41 (7%)
Query 104 DDKVLVDIGAGYMIQKTYQDEKKDAMRILTFMSEQQARLEK 144
D V ++ +GY IQ QD D+MR+L +M ++ A +
Sbjct 304 DGSVSSEVRSGYRIQANQQD---DSMRVLYYMEKELANFDP 341
Lambda K H
0.316 0.129 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4924747408
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40