bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2121_orf1
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
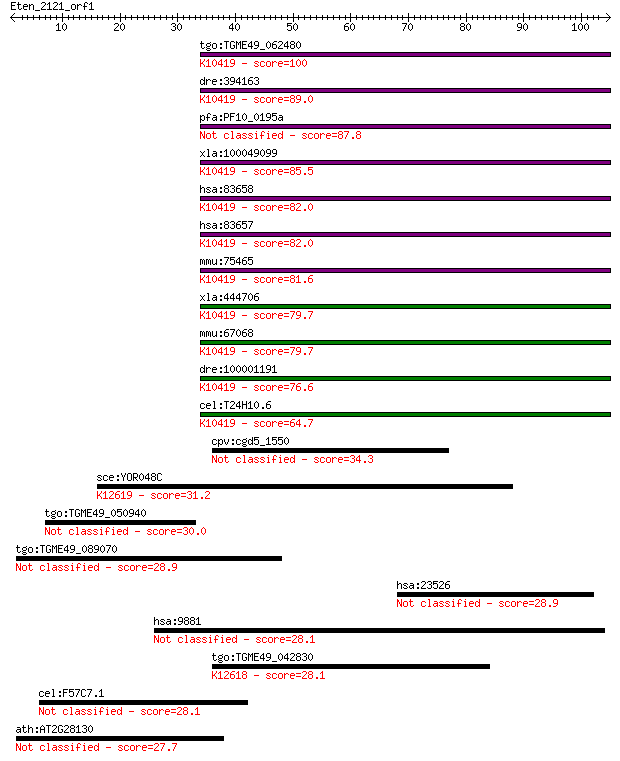
tgo:TGME49_062480 dynein light chain roadblock-type 2, putativ... 100 1e-21
dre:394163 dynlrb1, DNCL2A, MGC56335, zgc:56335; dynein, light... 89.0 3e-18
pfa:PF10_0195a flagellar outer arm dynein-associated protein, ... 87.8 7e-18
xla:100049099 dynlrb2; dynein, light chain, roadblock-type 2; ... 85.5 4e-17
hsa:83658 DYNLRB1, BITH, BLP, DNCL2A, DNLC2A, ROBLD1; dynein, ... 82.0 4e-16
hsa:83657 DYNLRB2, DNCL2B, DNLC2B, MGC62033, ROBLD2; dynein, l... 82.0 5e-16
mmu:75465 Dynlrb2, 1700009A04Rik, DNLC2B, Dncl2b; dynein light... 81.6 5e-16
xla:444706 dynlrb1, MGC84432; dynein, light chain, roadblock-t... 79.7 2e-15
mmu:67068 Dynlrb1, 2010012N15Rik, 2010320M17Rik, 9430076K19Rik... 79.7 2e-15
dre:100001191 dynein, light chain, roadblock-type 2-like; K104... 76.6 2e-14
cel:T24H10.6 dyrb-1; DYnein light chain (RoadBlock type) famil... 64.7 7e-11
cpv:cgd5_1550 hypothetical protein 34.3 0.091
sce:YOR048C RAT1, HKE1, TAP1, XRN2; Nuclear 5' to 3' single-st... 31.2 0.73
tgo:TGME49_050940 hypothetical protein 30.0 1.8
tgo:TGME49_089070 P-Type cation-transporting ATPase, putative ... 28.9 3.8
hsa:23526 HMHA1, HA-1, HLA-HA1, KIAA0223; histocompatibility (... 28.9 4.5
hsa:9881 TRANK1, KIAA0342, LBA1; tetratricopeptide repeat and ... 28.1 6.2
tgo:TGME49_042830 5'-3' exonuclease, putative ; K12618 5'-3' e... 28.1 6.3
cel:F57C7.1 hypothetical protein 28.1 7.4
ath:AT2G28130 hypothetical protein 27.7 8.2
> tgo:TGME49_062480 dynein light chain roadblock-type 2, putative
; K10419 dynein light chain roadblock-type
Length=102
Score = 100 bits (249), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 47/71 (66%), Positives = 59/71 (83%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
M+EVEETLNRIKT KGV GI+++ +DG +RSTL Q TA Y+A +QLA +ARS+VRDL
Sbjct 1 MSEVEETLNRIKTHKGVCGIVIVNSDGVPIRSTLDSQTTAQYSALISQLAAKARSVVRDL 60
Query 94 DPQNDVTFLRV 104
DPQND+TFLR+
Sbjct 61 DPQNDLTFLRI 71
> dre:394163 dynlrb1, DNCL2A, MGC56335, zgc:56335; dynein, light
chain, roadblock-type 1; K10419 dynein light chain roadblock-type
Length=96
Score = 89.0 bits (219), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 41/71 (57%), Positives = 55/71 (77%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
MAEVEET+ RI+++KGV+GII++ A+G ++STL T YAA QL +AR +VRD+
Sbjct 1 MAEVEETIKRIQSQKGVQGIIIVNAEGIPIKSTLDNTSTVQYAANIHQLLMKARGIVRDI 60
Query 94 DPQNDVTFLRV 104
DPQND+TFLRV
Sbjct 61 DPQNDLTFLRV 71
> pfa:PF10_0195a flagellar outer arm dynein-associated protein,
putative
Length=101
Score = 87.8 bits (216), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 37/71 (52%), Positives = 54/71 (76%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
M+E EE LNRIK KGV GI+V+ ++G I++ST QQQ+ +A+ QL+ +AR ++R+L
Sbjct 1 MSEAEEILNRIKNHKGVIGILVVNSEGLIIKSTFDQQQSDLHASLLTQLSKKARDVIREL 60
Query 94 DPQNDVTFLRV 104
DPQND+ FLR+
Sbjct 61 DPQNDINFLRL 71
> xla:100049099 dynlrb2; dynein, light chain, roadblock-type 2;
K10419 dynein light chain roadblock-type
Length=96
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 40/71 (56%), Positives = 52/71 (73%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
MAEVEETL RI++ KGV G IV+ A+G +R+TL T YA QL+ +A+S VRD+
Sbjct 1 MAEVEETLKRIQSHKGVIGTIVVNAEGIPIRTTLDNSTTVQYAGLLHQLSMKAKSTVRDI 60
Query 94 DPQNDVTFLRV 104
DPQND+TFLR+
Sbjct 61 DPQNDLTFLRI 71
> hsa:83658 DYNLRB1, BITH, BLP, DNCL2A, DNLC2A, ROBLD1; dynein,
light chain, roadblock-type 1; K10419 dynein light chain roadblock-type
Length=96
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 37/71 (52%), Positives = 52/71 (73%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
MAEVEETL R++++KGV+GIIV+ +G ++ST+ T YA+ +ARS VRD+
Sbjct 1 MAEVEETLKRLQSQKGVQGIIVVNTEGIPIKSTMDNPTTTQYASLMHSFILKARSTVRDI 60
Query 94 DPQNDVTFLRV 104
DPQND+TFLR+
Sbjct 61 DPQNDLTFLRI 71
> hsa:83657 DYNLRB2, DNCL2B, DNLC2B, MGC62033, ROBLD2; dynein,
light chain, roadblock-type 2; K10419 dynein light chain roadblock-type
Length=96
Score = 82.0 bits (201), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 38/71 (53%), Positives = 50/71 (70%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
MAEVEETL RI++ KGV G +V+ A+G +R+TL T YA L +A+S VRD+
Sbjct 1 MAEVEETLKRIQSHKGVIGTMVVNAEGIPIRTTLDNSTTVQYAGLLHHLTMKAKSTVRDI 60
Query 94 DPQNDVTFLRV 104
DPQND+TFLR+
Sbjct 61 DPQNDLTFLRI 71
> mmu:75465 Dynlrb2, 1700009A04Rik, DNLC2B, Dncl2b; dynein light
chain roadblock-type 2; K10419 dynein light chain roadblock-type
Length=96
Score = 81.6 bits (200), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 38/71 (53%), Positives = 50/71 (70%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
M EVEETL RI++ KGV G +V+ A+G +R+TL T YA QL +A+S VRD+
Sbjct 1 MTEVEETLKRIQSHKGVIGTMVVNAEGIPIRTTLDNSTTVQYAGLLHQLTMKAKSTVRDI 60
Query 94 DPQNDVTFLRV 104
DPQND+TFLR+
Sbjct 61 DPQNDLTFLRI 71
> xla:444706 dynlrb1, MGC84432; dynein, light chain, roadblock-type
1; K10419 dynein light chain roadblock-type
Length=96
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 37/71 (52%), Positives = 52/71 (73%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
MA+VEETL RI+ +KGV+GII++ ++G ++ST+ Q T YA QL +AR VRD+
Sbjct 1 MADVEETLKRIQGQKGVQGIIIVNSEGIPIKSTMDNQTTVQYAGLMHQLVLKARCSVRDI 60
Query 94 DPQNDVTFLRV 104
D QND+TFLR+
Sbjct 61 DAQNDLTFLRI 71
> mmu:67068 Dynlrb1, 2010012N15Rik, 2010320M17Rik, 9430076K19Rik,
AV124457, DNLC2A, Dncl2a, MGC159313, MGC159351; dynein light
chain roadblock-type 1; K10419 dynein light chain roadblock-type
Length=96
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 36/71 (50%), Positives = 51/71 (71%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
MAEVEETL R++++KGV+GIIV+ +G ++ST+ T YA +ARS VR++
Sbjct 1 MAEVEETLKRLQSQKGVQGIIVVNTEGIPIKSTMDNPTTTQYANLMHNFILKARSTVREI 60
Query 94 DPQNDVTFLRV 104
DPQND+TFLR+
Sbjct 61 DPQNDLTFLRI 71
> dre:100001191 dynein, light chain, roadblock-type 2-like; K10419
dynein light chain roadblock-type
Length=96
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 36/71 (50%), Positives = 49/71 (69%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
MAEVE+TL RI+ + GV G IV+ +G +R+TL + YA QL +ARS VRD+
Sbjct 1 MAEVEDTLKRIQAQHGVIGTIVVNGEGIPIRTTLDNSTSVQYAGLLHQLTMKARSAVRDI 60
Query 94 DPQNDVTFLRV 104
DPQN++TFLR+
Sbjct 61 DPQNELTFLRI 71
> cel:T24H10.6 dyrb-1; DYnein light chain (RoadBlock type) family
member (dyrb-1); K10419 dynein light chain roadblock-type
Length=95
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 49/71 (69%), Gaps = 0/71 (0%)
Query 34 MAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADRARSLVRDL 93
M++ EET+ R+++ KGV GIIV+ + G ++ ST+ T ++ A QL ++ ++ +R+L
Sbjct 1 MSDFEETIRRLQSEKGVVGIIVVDSAGRVIHSTIDSDATQSHTAFLQQLCEKTKTSIREL 60
Query 94 DPQNDVTFLRV 104
D ND+TFLR+
Sbjct 61 DSSNDLTFLRL 71
> cpv:cgd5_1550 hypothetical protein
Length=94
Score = 34.3 bits (77), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Query 36 EVEETLNRIKTRKGVKGIIVLAADGCI-LRSTLGQQQTAAYA 76
++ ETL+RIK +KGV GII+L +D L ST ++ + Y
Sbjct 2 DLNETLSRIKGKKGVVGIIILRSDNMTPLISTFDKKTSRNYT 43
> sce:YOR048C RAT1, HKE1, TAP1, XRN2; Nuclear 5' to 3' single-stranded
RNA exonuclease, involved in RNA metabolism, including
rRNA and snRNA processing as well as poly (A+) dependent
and independent mRNA transcription termination (EC:3.1.13.-);
K12619 5'-3' exoribonuclease 2 [EC:3.1.13.-]
Length=1006
Score = 31.2 bits (69), Expect = 0.73, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 35/73 (47%), Gaps = 2/73 (2%)
Query 16 PCCFLRSVTPVFAEASKKMAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAY 75
PC + P E +A E T NR+ + ++V+A DG R+ + QQ+ +
Sbjct 60 PCSHPENKPPPETEDEMLLAVFEYT-NRVLNMARPRKVLVMAVDGVAPRAKMNQQRARRF 118
Query 76 AAAA-AQLADRAR 87
+A AQ+ + AR
Sbjct 119 RSARDAQIENEAR 131
> tgo:TGME49_050940 hypothetical protein
Length=831
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 10/26 (38%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 7 PRVWLGPSTPCCFLRSVTPVFAEASK 32
PR+W+ PS+ CF ++ +P+F E +
Sbjct 506 PRLWVRPSSCSCFSKTKSPLFDEHNH 531
> tgo:TGME49_089070 P-Type cation-transporting ATPase, putative
(EC:3.6.3.1 3.6.3.8 1.14.12.10 3.6.3.9)
Length=1947
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 21/46 (45%), Gaps = 0/46 (0%)
Query 2 PQTLNPRVWLGPSTPCCFLRSVTPVFAEASKKMAEVEETLNRIKTR 47
P R L +T CF+R TP A S K +E L R++ R
Sbjct 226 PWFFRLRCPLSEATAVCFVREPTPCAASGSNKAGPSDEALPRLRER 271
> hsa:23526 HMHA1, HA-1, HLA-HA1, KIAA0223; histocompatibility
(minor) HA-1
Length=1136
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 68 GQQQTAAYAAAAAQLADRARSLVRDLDPQNDVTF 101
G+Q + A A LA R R L+RDL P+N +
Sbjct 873 GRQDGSESEAVAVALAGRLRELLRDLPPENRASL 906
> hsa:9881 TRANK1, KIAA0342, LBA1; tetratricopeptide repeat and
ankyrin repeat containing 1
Length=2925
Score = 28.1 bits (61), Expect = 6.2, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 41/79 (51%), Gaps = 6/79 (7%)
Query 26 VFAEASKKMAEVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAAAAQLADR 85
VF ++ K++AE + LNR R+ + +++ GC+L + + TA A+ L
Sbjct 1897 VFLKSRKRLAEAADLLNREGRRE--EAALLMKQHGCLLEAA---RLTADKDFQASCLLGA 1951
Query 86 AR-SLVRDLDPQNDVTFLR 103
AR ++ RD D ++ LR
Sbjct 1952 ARLNVARDSDIEHTKDILR 1970
> tgo:TGME49_042830 5'-3' exonuclease, putative ; K12618 5'-3'
exoribonuclease 1 [EC:3.1.13.-]
Length=2042
Score = 28.1 bits (61), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 28/50 (56%), Gaps = 2/50 (4%)
Query 36 EVEETLNRIKTRKGVKGIIVLAADGCILRSTLGQQQTAAYAAA--AAQLA 83
+V L+ I + K +VLAADG R+ + QQ+ Y AA AA+LA
Sbjct 62 DVFAALDLIISTVNPKKFLVLAADGVAPRAKMNQQRARRYRAAKDAAELA 111
> cel:F57C7.1 hypothetical protein
Length=1209
Score = 28.1 bits (61), Expect = 7.4, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 6 NPRVWLGPSTPCCFLRSVTPVFAEASKKMAEVEETL 41
+P+ G +TPC +PV E + KM EVEE +
Sbjct 153 SPQKPQGSTTPCSVQSPASPVGDEQAVKMEEVEEPI 188
> ath:AT2G28130 hypothetical protein
Length=458
Score = 27.7 bits (60), Expect = 8.2, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 20/37 (54%), Gaps = 3/37 (8%)
Query 2 PQTLNPRVWLGPSTPCCFLRSVTPVFAEAS-KKMAEV 37
PQ N R WL T CC R P+F + +++AE+
Sbjct 248 PQ--NIRAWLTLVTTCCQFRCKKPIFTTSQVEQIAEI 282
Lambda K H
0.320 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2022291452
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40