bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2148_orf1
Length=340
Score E
Sequences producing significant alignments: (Bits) Value
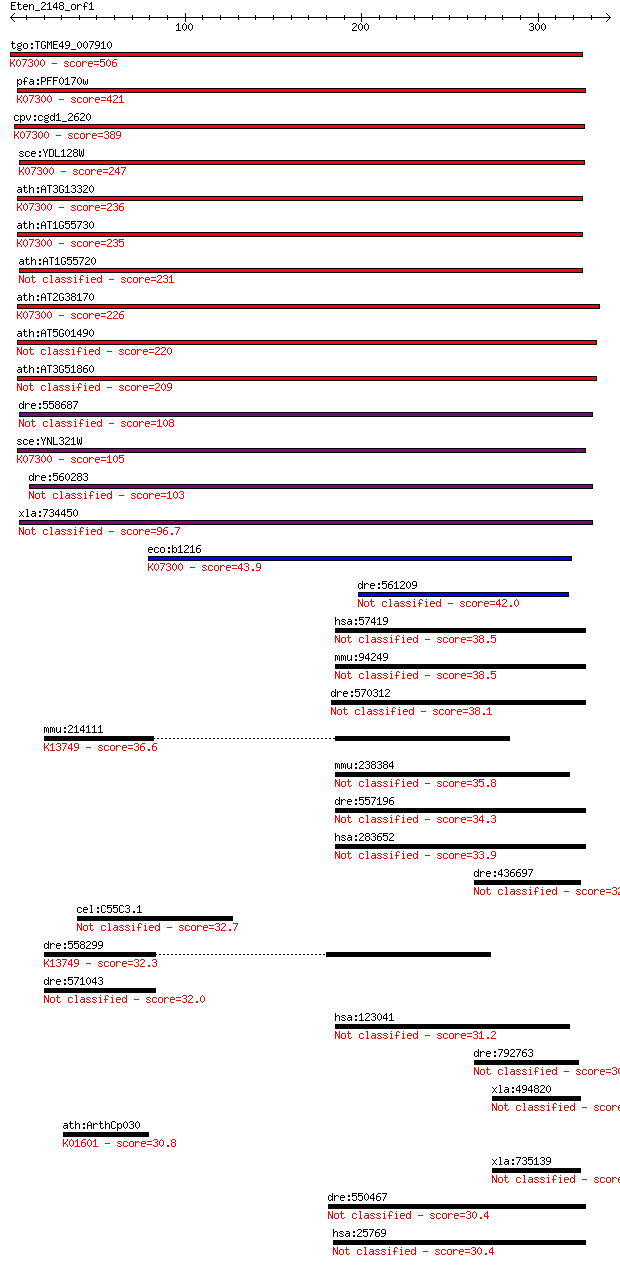
tgo:TGME49_007910 manganese resistance 1 protein, putative ; K... 506 5e-143
pfa:PFF0170w calcium antiporter, putative; K07300 Ca2+:H+ anti... 421 3e-117
cpv:cgd1_2620 calcium antiporter, Na/Ca exchange associated me... 389 9e-108
sce:YDL128W VCX1, HUM1, MNR1; Vacuolar H+/Ca2+ exchanger invol... 247 5e-65
ath:AT3G13320 CAX2; CAX2 (CATION EXCHANGER 2); calcium:cation ... 236 1e-61
ath:AT1G55730 ATCAX5; ATCAX5; calcium:cation antiporter/ catio... 235 2e-61
ath:AT1G55720 ATCAX6; ATCAX6; calcium:cation antiporter/ catio... 231 4e-60
ath:AT2G38170 CAX1; CAX1 (cation exchanger 1); calcium ion tra... 226 1e-58
ath:AT5G01490 CAX4; CAX4 (CATION EXCHANGER 4); calcium:cation ... 220 6e-57
ath:AT3G51860 CAX3; CAX3 (CATION EXCHANGER 3); calcium:cation ... 209 1e-53
dre:558687 type II CAX cation/proton exchanger-like 108 4e-23
sce:YNL321W VNX1; Low affinity vacuolar membrane localized mon... 105 3e-22
dre:560283 cax1, MGC136271, si:dkey-180p18.2, wu:fc25f10, zgc:... 103 9e-22
xla:734450 hypothetical protein MGC115264 96.7 1e-19
eco:b1216 chaA, ECK1210, JW1207; calcium/sodium:proton antipor... 43.9 8e-04
dre:561209 solute carrier family 24 (sodium/potassium/calcium ... 42.0 0.004
hsa:57419 SLC24A3, NCKX3; solute carrier family 24 (sodium/pot... 38.5 0.035
mmu:94249 Slc24a3, NCKX3; solute carrier family 24 (sodium/pot... 38.5 0.038
dre:570312 slc24a5; solute carrier family 24, member 5; K13753... 38.1 0.043
mmu:214111 Slc24a1, MGC27617; solute carrier family 24 (sodium... 36.6 0.13
mmu:238384 Slc24a4, A930002M03Rik, AW492432, NCKX4; solute car... 35.8 0.27
dre:557196 slc24a3, si:dkey-187a12.3; solute carrier family 24... 34.3 0.67
hsa:283652 SLC24A5, JSX, NCKX5, SHEP4; solute carrier family 2... 33.9 1.0
dre:436697 slc48a1a, zgc:92662; solute carrier family 48 (heme... 32.7 1.8
cel:C55C3.1 hypothetical protein 32.7 2.2
dre:558299 slc24a1, si:ch211-117i10.3; solute carrier family 2... 32.3 2.7
dre:571043 NCKX2.3-like 32.0 3.7
hsa:123041 SLC24A4, FLJ38852, NCKX4, SHEP6, SLC24A2; solute ca... 31.2 6.6
dre:792763 slc48a1b, MGC174054, MGC63994, fj61c11, hrg1, wu:fj... 30.8 8.2
xla:494820 slc48a1-a, HRG-1A, hHRG-1, hrg1, hrg1-a, slc48a1; s... 30.8 8.4
ath:ArthCp030 rbcL; RuBisCO large subunit; K01601 ribulose-bis... 30.8 8.4
xla:735139 slc48a1-b, HRG-1B, MGC115395, hHRG-1, hrg1, hrg1-b;... 30.4 8.9
dre:550467 zgc:112072; K13752 solute carrier family 24 (sodium... 30.4 9.4
hsa:25769 SLC24A2, FLJ45149, NCKX2; solute carrier family 24 (... 30.4 9.9
> tgo:TGME49_007910 manganese resistance 1 protein, putative ;
K07300 Ca2+:H+ antiporter
Length=477
Score = 506 bits (1303), Expect = 5e-143, Method: Compositional matrix adjust.
Identities = 255/325 (78%), Positives = 290/325 (89%), Gaps = 1/325 (0%)
Query 1 STLCVFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGL 60
S L VF NF ALIPL++LLG TEEL+LHTGEI GGLLNATFGNAVEMIMSVQALR+GL
Sbjct 153 SPLFVFICNFLALIPLASLLGNATEELALHTGEIIGGLLNATFGNAVEMIMSVQALRVGL 212
Query 61 LEVVKGTLLGSILSNLLLVLGMSFFAGGLHHYLQKFNEKGATCSVTLLLLSCMGIVIPTV 120
L VV+GT+LGSILSNLLLVLGMSFFAGG+ +++QKFNEKGATCSVTLLLLSCM IVIPTV
Sbjct 213 LSVVQGTMLGSILSNLLLVLGMSFFAGGIRYHVQKFNEKGATCSVTLLLLSCMSIVIPTV 272
Query 121 AAVDNGQ-HGTYNILMISRITALLIGVTYCLFLFFQLYTHIGLFKDDDEDAEQWPMMSWE 179
AA N + TY+I+ ISR A+LIGVTYCLFLFFQLYTH+ LF+DD+E E WP MSWE
Sbjct 273 AASGNDHANPTYDIIKISRTIAVLIGVTYCLFLFFQLYTHLNLFRDDEEGEEGWPSMSWE 332
Query 180 AATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAM 239
AAT+MLF+VT L+A+HSE LV SI +VV +YGL ESFIGVILLPIVGNAAEHLTAVTVAM
Sbjct 333 AATVMLFIVTLLIAVHSEYLVGSIHDVVTNYGLPESFIGVILLPIVGNAAEHLTAVTVAM 392
Query 240 KNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSVLVAMAVV 299
KNKVDLAMGVAVGSSAQIALFVFPFTV WV+ QPLT+AVQP++A+V+L++VLVAMA+V
Sbjct 393 KNKVDLAMGVAVGSSAQIALFVFPFTVCAGWVLDQPLTLAVQPMNALVLLMAVLVAMAIV 452
Query 300 QDGESNWLEGVMLMAAYLIISVVFW 324
QDGESNWLEGVMLMAAYL+I++VFW
Sbjct 453 QDGESNWLEGVMLMAAYLMIAIVFW 477
> pfa:PFF0170w calcium antiporter, putative; K07300 Ca2+:H+ antiporter
Length=441
Score = 421 bits (1081), Expect = 3e-117, Method: Compositional matrix adjust.
Identities = 216/322 (67%), Positives = 264/322 (81%), Gaps = 2/322 (0%)
Query 5 VFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVV 64
+F FNF LIPLSAL+G TE+L+LHTGEI GGLLNATFGN +EMI S+QAL GL+ VV
Sbjct 120 IFFFNFMVLIPLSALMGHVTEDLALHTGEIIGGLLNATFGNLMEMIFSIQALNAGLINVV 179
Query 65 KGTLLGSILSNLLLVLGMSFFAGGLHHYLQKFNEKGATCSVTLLLLSCMGIVIPTVAAVD 124
+GTLLGSILSNLLLVLGMSFFAGGL+H++QKFNEKGATCS +LLLLS + I IPTV++
Sbjct 180 QGTLLGSILSNLLLVLGMSFFAGGLYHHIQKFNEKGATCSTSLLLLSSLAITIPTVSSFT 239
Query 125 NGQHGTYNILMISRITALLIGVTYCLFLFFQLYTHIGLFKDDDEDAEQWPMMSWEAATLM 184
+ IL +SRITA+LI VTYCLFL FQLYTHI LF+D E E+ P +S + ++
Sbjct 240 TNNNLDV-ILKVSRITAVLIFVTYCLFLLFQLYTHISLFQDK-EMTEEIPQLSVISGSIF 297
Query 185 LFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVD 244
L L+T LV++HSE L+ SI+ V+ Y +SE+FIGVILLP+VGNA EHLTAVTVAMKNKVD
Sbjct 298 LILITLLVSIHSEFLIYSIDSVIKYYNISENFIGVILLPVVGNATEHLTAVTVAMKNKVD 357
Query 245 LAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSVLVAMAVVQDGES 304
L MGVAVGSSAQIALFV P TVL W++ +P+T+A PLS V++++SV+V MA+VQDGES
Sbjct 358 LTMGVAVGSSAQIALFVVPVTVLFGWILNKPMTLAFSPLSTVILVISVIVTMAIVQDGES 417
Query 305 NWLEGVMLMAAYLIISVVFWFD 326
NWLEGV+L++AYLI+ VVFWFD
Sbjct 418 NWLEGVLLISAYLIVGVVFWFD 439
> cpv:cgd1_2620 calcium antiporter, Na/Ca exchange associated
membrane protein with 11 transmembrane domains ; K07300 Ca2+:H+
antiporter
Length=473
Score = 389 bits (999), Expect = 9e-108, Method: Compositional matrix adjust.
Identities = 201/376 (53%), Positives = 265/376 (70%), Gaps = 56/376 (14%)
Query 3 LCVFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLE 62
L F FNF ALIPL+ LLG FTEEL+LHTGE+ GGLLNATFGNAVE I++VQ +R GL+
Sbjct 100 LATFWFNFIALIPLANLLGIFTEELALHTGEVVGGLLNATFGNAVEAILTVQGIRAGLIT 159
Query 63 VVKGTLLGSILSNLLLVLGMSFFAGGLHHYLQKFNEKGATCSVTLLLLSCMGIVIPT-VA 121
VV+GTLLGSILSNLLLVLGMSFFAGG+ H++QKFNEKGA+ S +LL+LSCM I IPT VA
Sbjct 160 VVQGTLLGSILSNLLLVLGMSFFAGGIFHHVQKFNEKGASFSTSLLMLSCMAISIPTIVA 219
Query 122 AVDNGQHGTYNILMISRITALLIGVTYCLFLFFQLYTHIGLFKD-----DDEDAEQW--- 173
D QH NILMISR+TA+L+ TY LFLFFQLYTHI LF+D +D+ +
Sbjct 220 QFDLPQH---NILMISRLTAILLSFTYVLFLFFQLYTHINLFRDESVASNDKSYPSYMID 276
Query 174 --------------------------------------------PMMSWEAATLMLFLVT 189
P +SW+ T+++ L T
Sbjct 277 NISSVKYPNALPNIEIYNNNIYKNYPIISQNFNNYQLYDVCLELPTISWQIGTVLILLCT 336
Query 190 SLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVDLAMGV 249
L+++ SE LV SI ++++ SE+FIGVILLP+VGNAAEH+TAV+VA+KNK DL +GV
Sbjct 337 ILISIISECLVDSINGFISEWRFSENFIGVILLPLVGNAAEHITAVSVAIKNKTDLTIGV 396
Query 250 AVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSVLVAMAVVQDGESNWLEG 309
A+GSS QIALFV PF+V+V W++G+P+T++ P+SA+++LL+ L+ + +VQDGESNW EG
Sbjct 397 AIGSSTQIALFVVPFSVIVGWLLGKPMTLSFTPVSAIILLLTNLIVIGIVQDGESNWFEG 456
Query 310 VMLMAAYLIISVVFWF 325
++L+ +Y I++VV+W+
Sbjct 457 ILLIISYCIVAVVYWY 472
> sce:YDL128W VCX1, HUM1, MNR1; Vacuolar H+/Ca2+ exchanger involved
in control of cytosolic Ca2+ concentration; has similarity
to sodium/calcium exchangers, including the bovine Na+/Ca2+,K+
antiporter; K07300 Ca2+:H+ antiporter
Length=411
Score = 247 bits (630), Expect = 5e-65, Method: Compositional matrix adjust.
Identities = 146/334 (43%), Positives = 227/334 (67%), Gaps = 14/334 (4%)
Query 6 FSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVVK 65
F FNF A+IPL+A+L TEEL+ G GGLLNATFGNAVE+I+S+ AL+ G + +V+
Sbjct 64 FLFNFLAIIPLAAILANATEELADKAGNTIGGLLNATFGNAVELIVSIIALKKGQVRIVQ 123
Query 66 GTLLGSILSNLLLVLGMSFFAGGLHHYLQKFNEKGATCSVTLLLLSCMGIVIPTV--AAV 123
++LGS+LSNLLLVLG+ F GG + Q FN+ A +LL ++C ++IP A +
Sbjct 124 ASMLGSLLSNLLLVLGLCFIFGGYNRVQQTFNQTAAQTMSSLLAIACASLLIPAAFRATL 183
Query 124 DNGQHGTY---NILMISRITALLIGVTYCLFLFFQLYTHIGLFKDDDEDAEQ-------- 172
+G+ + IL +SR T+++I + Y LFL+FQL +H LF+ +E+ ++
Sbjct 184 PHGKEDHFIDGKILELSRGTSIVILIVYVLFLYFQLGSHHALFEQQEEETDEVMSTISRN 243
Query 173 -WPMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEH 231
+S +++ ++L T +++ ++ LV +I+ VV GLS++FIG+I++PIVGNAAEH
Sbjct 244 PHHSLSVKSSLVILLGTTVIISFCADFLVGTIDNVVESTGLSKTFIGLIVIPIVGNAAEH 303
Query 232 LTAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLS 291
+T+V VAMK+K+DLA+GVA+GSS Q+ALFV PF VLV W++ P+T+ + ++
Sbjct 304 VTSVLVAMKDKMDLALGVAIGSSLQVALFVTPFMVLVGWMIDVPMTLNFSTFETATLFIA 363
Query 292 VLVAMAVVQDGESNWLEGVMLMAAYLIISVVFWF 325
V ++ ++ DGESNWLEGVM +A Y++I++ F++
Sbjct 364 VFLSNYLILDGESNWLEGVMSLAMYILIAMAFFY 397
> ath:AT3G13320 CAX2; CAX2 (CATION EXCHANGER 2); calcium:cation
antiporter/ calcium:hydrogen antiporter; K07300 Ca2+:H+ antiporter
Length=441
Score = 236 bits (602), Expect = 1e-61, Method: Compositional matrix adjust.
Identities = 137/332 (41%), Positives = 208/332 (62%), Gaps = 12/332 (3%)
Query 5 VFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVV 64
VF + PL+ LG TE+L+ +TG GGLLNATFGN E+I+S+ AL+ G++ VV
Sbjct 100 VFLLTLVGITPLAERLGYATEQLACYTGPTVGGLLNATFGNVTELIISIFALKNGMIRVV 159
Query 65 KGTLLGSILSNLLLVLGMSFFAGGLHHYL--QKFNEKGATCSVTLLLLSCMGIVIPTVAA 122
+ TLLGSILSN+LLVLG +FF GGL Y Q F++ AT + LLL++ MGI+ P V
Sbjct 160 QLTLLGSILSNMLLVLGCAFFCGGLVFYQKDQVFDKGIATVNSGLLLMAVMGILFPAVLH 219
Query 123 VDNGQ-HGTYNILMISRITALLIGVTYCLFLFFQLYTHIGLFKDDDEDAEQW-------- 173
+ + H + L +SR ++ ++ + Y +LFFQL + + DE++ Q
Sbjct 220 YTHSEVHAGSSELALSRFSSCIMLIAYAAYLFFQLKSQSNSYSPLDEESNQNEETSAEDE 279
Query 174 -PMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHL 232
P +S A + L ++T+ V+L S LV +IE + + +FI ILLPIVGNAAEH
Sbjct 280 DPEISKWEAIIWLSILTAWVSLLSGYLVDAIEGASVSWNIPIAFISTILLPIVGNAAEHA 339
Query 233 TAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSV 292
A+ AMK+K+DL++GVA+GSS QI++F PF V++ W+MGQ + + Q ++ ++V
Sbjct 340 GAIMFAMKDKLDLSLGVAIGSSIQISMFAVPFCVVIGWMMGQQMDLNFQLFETAMLFITV 399
Query 293 LVAMAVVQDGESNWLEGVMLMAAYLIISVVFW 324
+V +Q+G SN+ +G+ML+ YLI++ F+
Sbjct 400 IVVAFFLQEGSSNYFKGLMLILCYLIVAASFF 431
> ath:AT1G55730 ATCAX5; ATCAX5; calcium:cation antiporter/ cation:cation
antiporter; K07300 Ca2+:H+ antiporter
Length=441
Score = 235 bits (600), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 138/336 (41%), Positives = 207/336 (61%), Gaps = 20/336 (5%)
Query 5 VFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVV 64
+F + + PL+ LG TE+L+ +TG GGLLNATFGN E+I+S+ AL+ G++ VV
Sbjct 100 IFLLSLVGITPLAERLGYATEQLACYTGSTVGGLLNATFGNVTELIISIFALKSGMIRVV 159
Query 65 KGTLLGSILSNLLLVLGMSFFAGGL--HHYLQKFNEKGATCSVTLLLLSCMGIVIPTVAA 122
+ TLLGSILSN+LLVLG +FF GGL Q F++ A + LLL++ MG++ P V
Sbjct 160 QLTLLGSILSNMLLVLGCAFFCGGLVFSQKEQVFDKGNAVVNSGLLLMAVMGLLFPAVLH 219
Query 123 VDNGQ-HGTYNILMISRITALLIGVTYCLFLFFQLYTHIGLF-------------KDDDE 168
+ + H + L +SR ++ ++ V Y +LFFQL + + DDDE
Sbjct 220 YTHSEVHAGSSELALSRFSSCIMLVAYAAYLFFQLKSQPSSYTPLTEETNQNEETSDDDE 279
Query 169 DAEQWPMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNA 228
D E + WEA + L ++T+ V+L S LV +IE + + SFI VILLPIVGNA
Sbjct 280 DPE---ISKWEA-IIWLSILTAWVSLLSGYLVDAIEGASVSWKIPISFISVILLPIVGNA 335
Query 229 AEHLTAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVM 288
AEH A+ AMK+K+DL++GVA+GSS QI++F PF V++ W+MG + + Q +
Sbjct 336 AEHAGAIMFAMKDKLDLSLGVAIGSSIQISMFAVPFCVVIGWMMGAQMDLNFQLFETATL 395
Query 289 LLSVLVAMAVVQDGESNWLEGVMLMAAYLIISVVFW 324
++V+V +Q+G SN+ +G+ML+ YLI++ F+
Sbjct 396 FITVIVVAFFLQEGTSNYFKGLMLILCYLIVAASFF 431
> ath:AT1G55720 ATCAX6; ATCAX6; calcium:cation antiporter/ cation:cation
antiporter
Length=467
Score = 231 bits (588), Expect = 4e-60, Method: Compositional matrix adjust.
Identities = 138/333 (41%), Positives = 207/333 (62%), Gaps = 18/333 (5%)
Query 6 FSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVVK 65
F + + PL+ LG TE+LS +TG GGLLNATFGN +E+I+S+ AL+ G++ VV+
Sbjct 112 FLLSLVGITPLAERLGYATEQLSCYTGATVGGLLNATFGNVIELIISIIALKNGMIRVVQ 171
Query 66 GTLLGSILSNLLLVLGMSFFAGGL--HHYLQKFNEKGATCSVTLLLLSCMGIVIPTVAAV 123
TLLGSILSN+LLVLG +FF GGL Q F+++ A S +LL++ MG++ PT
Sbjct 172 LTLLGSILSNILLVLGCAFFCGGLVFPGKDQVFDKRNAVVSSGMLLMAVMGLLFPTFLHY 231
Query 124 DNGQ-HGTYNILMISRITALLIGVTYCLFLFFQLYTHIGLF----------KDDDEDAEQ 172
+ + H + L +SR + ++ V Y +LFFQL + + +DDED E
Sbjct 232 THSEVHAGSSELALSRFISCIMLVAYAAYLFFQLKSQPSFYTEKTNQNEETSNDDEDPE- 290
Query 173 WPMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHL 232
+ WEA + L + T+ V+L S LV +IE + + SFI VILLPIVGNAAEH
Sbjct 291 --ISKWEA-IIWLSIFTAWVSLLSGYLVDAIEGTSVSWKIPISFISVILLPIVGNAAEHA 347
Query 233 TAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSV 292
A+ AMK+K+DL++GVA+GSS QI++F PF V++ W+MG + + +Q +L++V
Sbjct 348 GAIMFAMKDKLDLSLGVAIGSSIQISMFAVPFCVVIGWMMGAQMDLNLQLFETATLLITV 407
Query 293 LVAMAVVQ-DGESNWLEGVMLMAAYLIISVVFW 324
+V +Q +G SN+ + +ML+ YLI++ F+
Sbjct 408 IVVAFFLQLEGTSNYFKRLMLILCYLIVAASFF 440
> ath:AT2G38170 CAX1; CAX1 (cation exchanger 1); calcium ion transmembrane
transporter/ calcium:cation antiporter/ calcium:hydrogen
antiporter; K07300 Ca2+:H+ antiporter
Length=463
Score = 226 bits (575), Expect = 1e-58, Method: Compositional matrix adjust.
Identities = 143/351 (40%), Positives = 218/351 (62%), Gaps = 27/351 (7%)
Query 5 VFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVV 64
+F + L PL+ + TE+L+ +TG GGLLNAT GNA E+I+++ AL + VV
Sbjct 98 IFGLSLLGLTPLAERVSFLTEQLAFYTGPTLGGLLNATCGNATELIIAILALTNNKVAVV 157
Query 65 KGTLLGSILSNLLLVLGMSFFAGGLHHYL--QKFNEKGATCSVTLLLLSCMGIVIPT-VA 121
K +LLGSILSNLLLVLG S F GG+ + Q+F+ K A + LLLL + ++P V
Sbjct 158 KYSLLGSILSNLLLVLGTSLFCGGIANIRREQRFDRKQADVNFFLLLLGFLCHLLPLLVG 217
Query 122 AVDNGQHGTYNI----LMISRITALLIGVTYCLFLFFQLYTHIGLF----KDD--DEDAE 171
+ NG+ + L ISR ++++ ++Y +L FQL+TH LF ++D D+D E
Sbjct 218 YLKNGEASAAVLSDMQLSISRGFSIVMLISYIAYLVFQLWTHRQLFDAQEQEDEYDDDVE 277
Query 172 Q-------WPMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPI 224
Q W +W L +T ++AL SE +V++IEE + LS SFI +ILLPI
Sbjct 278 QETAVISFWSGFAW------LVGMTLVIALLSEYVVATIEEASDKWNLSVSFISIILLPI 331
Query 225 VGNAAEHLTAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLS 284
VGNAAEH AV A KNK+D+++GVA+GS+ QI LFV P T++VAW++G + + PL
Sbjct 332 VGNAAEHAGAVIFAFKNKLDISLGVALGSATQIGLFVVPLTIIVAWILGINMDLNFGPLE 391
Query 285 AVVMLLSVLVAMAVVQDGESNWLEGVMLMAAYLIISVVFWFDK-PGPESAV 334
+ +S+++ +QDG S++++G++L+ Y II++ F+ DK P ++A+
Sbjct 392 TGCLAVSIIITAFTLQDGSSHYMKGLVLLLCYFIIAICFFVDKLPQKQNAI 442
> ath:AT5G01490 CAX4; CAX4 (CATION EXCHANGER 4); calcium:cation
antiporter/ calcium:hydrogen antiporter/ cation:cation antiporter
Length=454
Score = 220 bits (561), Expect = 6e-57, Method: Compositional matrix adjust.
Identities = 133/352 (37%), Positives = 206/352 (58%), Gaps = 30/352 (8%)
Query 5 VFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVV 64
VF+ + L PL+ + TE+++ HTG GGL+NAT GNA EMI+++ A+ + +V
Sbjct 99 VFALSLLGLTPLAERISFLTEQIAFHTGPTVGGLMNATCGNATEMIIAILAVGQRKMRIV 158
Query 65 KGTLLGSILSNLLLVLGMSFFAGGLHHYL--QKFNEKGATCSVTLLLLSCMGIVIPTVA- 121
K +LLGSILSNLL VLG S F GG+ + Q F+ + + LL L+ + +P +
Sbjct 159 KLSLLGSILSNLLFVLGTSLFLGGISNLRKHQSFDPRQGDMNSMLLYLALLCQTLPMIMR 218
Query 122 -AVDNGQHGTYNILMISRITALLIGVTYCLFLFFQLY------------THIGLFKDDDE 168
++ ++ +++++SR ++ ++ + Y FL F L+ + DD
Sbjct 219 FTMEAEEYDGSDVVVLSRASSFVMLIAYLAFLIFHLFSSHLSPPPPPLPQREDVHDDDVS 278
Query 169 DAEQ-------WPMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVIL 221
D E+ W + W L ++T LVAL S+ LVS+I++ +GLS FIG+IL
Sbjct 279 DKEEEGAVIGMWSAIFW------LIIMTLLVALLSDYLVSTIQDAADSWGLSVGFIGIIL 332
Query 222 LPIVGNAAEHLTAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQ 281
LPIVGNAAEH AV A +NK+D+ +G+A+GS+ QIALFV P TVLVAW MG + +
Sbjct 333 LPIVGNAAEHAGAVIFAFRNKLDITLGIALGSATQIALFVVPVTVLVAWTMGIEMDLNFN 392
Query 282 PLSAVVMLLSVLVAMAVVQDGESNWLEGVMLMAAYLIISVVFWF-DKPGPES 332
L LS+LV V+QDG SN+++G++L+ Y++I+ F+ + P E+
Sbjct 393 LLETACFALSILVTSLVLQDGTSNYMKGLVLLLCYVVIAACFFVSNSPSTET 444
> ath:AT3G51860 CAX3; CAX3 (CATION EXCHANGER 3); calcium:cation
antiporter/ calcium:hydrogen antiporter/ cation:cation antiporter
Length=459
Score = 209 bits (533), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 131/345 (37%), Positives = 209/345 (60%), Gaps = 17/345 (4%)
Query 5 VFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVV 64
+F + L PL+ + TE+L+ +TG GGLLNAT GNA E+I+++ AL + VV
Sbjct 98 IFGLSLIGLTPLAERVSFLTEQLAFYTGPTVGGLLNATCGNATELIIAILALANNKVAVV 157
Query 65 KGTLLGSILSNLLLVLGMSFFAGGLHHYL--QKFNEKGATCSV-TLLLLSCMGIVIPTVA 121
K +LLGSILSNLLLVLG S F GG+ + Q+F+ K A + LL+ ++ +
Sbjct 158 KYSLLGSILSNLLLVLGTSLFFGGIANIRREQRFDRKQADVNFFLLLMGLLCHLLPLLLK 217
Query 122 AVDNGQHGTYNI----LMISRITALLIGVTYCLFLFFQLYTHIGLFKDD---------DE 168
G+ T I L +SR +++++ + Y +L FQL+TH LF+ +
Sbjct 218 YAATGEVSTSMINKMSLTLSRTSSIVMLIAYIAYLIFQLWTHRQLFEAQQDDDDAYDDEV 277
Query 169 DAEQWPMMSWEAATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNA 228
E+ P++ + + L +T ++AL SE +V +IE+ +GLS SFI +ILLPIVGNA
Sbjct 278 SVEETPVIGFWSGFAWLVGMTIVIALLSEYVVDTIEDASDSWGLSVSFISIILLPIVGNA 337
Query 229 AEHLTAVTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVM 288
AEH A+ A KNK+D+++GVA+GS+ QI+LFV P +V+VAW++G + + L +
Sbjct 338 AEHAGAIIFAFKNKLDISLGVALGSATQISLFVVPLSVIVAWILGIKMDLNFNILETSSL 397
Query 289 LLSVLVAMAVVQDGESNWLEGVMLMAAYLIISVVFWFDK-PGPES 332
L++++ +QDG S++++G++L+ Y+II+ F+ D+ P P
Sbjct 398 ALAIIITAFTLQDGTSHYMKGLVLLLCYVIIAACFFVDQIPQPND 442
> dre:558687 type II CAX cation/proton exchanger-like
Length=746
Score = 108 bits (269), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 97/373 (26%), Positives = 169/373 (45%), Gaps = 52/373 (13%)
Query 6 FSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIG------ 59
F+ +++IPLS +G +S + G ++NATFG+ E+ + AL G
Sbjct 375 FATAVSSIIPLSYYIGMGIASISAQSNFAVGAVVNATFGSITELTFYISALLRGHHQGNK 434
Query 60 -LLEVVKGTLLGSILSNLLLVLGMSFFAGGLHHYLQKFNEKGATCSVTLLLLSCMGIVIP 118
L E+VK L G++L +L + G+ GGL H Q+FN + A S +LL +S G+ P
Sbjct 435 CLQEIVKAALTGTLLGCILFIPGICMIIGGLKHQEQRFNSRSAGVSSSLLFISIGGVFAP 494
Query 119 TVAA-------VDNGQHGT--------------YNI------LMISRITAL------LIG 145
T+ + +N + T Y++ L S I L L+
Sbjct 495 TLFSKAYGNLVCENCTNSTGTNSSGSFMCKNCHYDLSENNYSLFHSHIEPLVYTVSFLLP 554
Query 146 VTYCLFLFFQLYTHIGLFKDDDED--------AEQWPMMSWEAATLMLFLVTSLVALHSE 197
V+Y + L F L TH ++ D + W W A +++F T L++ ++
Sbjct 555 VSYIIGLIFTLKTHSHIYDIHIGDGMASHLGASVHWS--RWRALLILIF-ATVLMSACAD 611
Query 198 LLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVDLAMGVAVGSSAQI 257
L I+ +++ +S+ FIGV LL +V E + V A++NK+ L++ V + Q+
Sbjct 612 LTTEHIQPILSHSSVSQYFIGVTLLAMVPEIPEIVNGVQFALQNKISLSLEVGSCIAVQV 671
Query 258 ALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSVLVAMAVVQDGESNWLEGVMLMAAYL 317
+ P VL + L + SV++ + DG+ ++ +G L+ YL
Sbjct 672 CMIQIPVLVLFNVFYDVGFVLVFSDLHLWASIFSVILVNYIFMDGKCDYFQGTALVVVYL 731
Query 318 IISVVFWFDKPGP 330
I+ +++F P P
Sbjct 732 ILLALYFF-APSP 743
> sce:YNL321W VNX1; Low affinity vacuolar membrane localized monovalent
cation/H+ antiporter; member of the calcium exchanger
(CAX) family; potential Cdc28p substrate; K07300 Ca2+:H+
antiporter
Length=908
Score = 105 bits (261), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 93/373 (24%), Positives = 170/373 (45%), Gaps = 51/373 (13%)
Query 5 VFSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVV 64
+F + IPL+ +G +S T G ++NA F VE+ + AL+ +V
Sbjct 531 IFILCLTSTIPLAFYIGQAVASISAQTSMGVGAVINAFFSTIVEIFLYCVALQQKKGLLV 590
Query 65 KGTLLGSILSNLLLVLGMSFFAGGLHHYLQKFNEKGATCSVTLLLLSCMGIVIPTV---- 120
+G+++GSIL +LL+ G+S G L+ Q++N A S LL+ S + + +PTV
Sbjct 591 EGSMIGSILGAVLLLPGLSMCGGALNRKTQRYNPASAGVSSALLIFSMIVMFVPTVLYEI 650
Query 121 ----------AAVDNGQHGTYNILMISRI-----------TALLIGVTYCLFLFFQLYTH 159
A D ++ L +R+ A+++ Y + L+F L TH
Sbjct 651 YGGYSVNCADGANDRDCTFSHPPLKFNRLFTHVIQPMSISCAIVLFCAYIIGLWFTLRTH 710
Query 160 IGLF-------------KDDDEDAEQWPMMSWEAATLMLFLVTSLVALHSELLVSSIEEV 206
+ + ++++ P S +T +L + T L A+ +E+LVS ++ V
Sbjct 711 AKMIWQLPIADPTSTAPEQQEQNSHDAPNWSRSKSTCILLMSTLLYAIIAEILVSCVDAV 770
Query 207 VADY-GLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVDLAMGVAVGSSAQIALFVFP-- 263
+ D L+ F+G+ + ++ N E L A++ A+ V L+M + + Q+ L P
Sbjct 771 LEDIPSLNPKFLGLTIFALIPNTTEFLNAISFAIHGNVALSMEIGSAYALQVCLLQIPSL 830
Query 264 --FTVLVAWVMGQPL-TMAVQPLSAVVMLLSVLVAMAVV-------QDGESNWLEGVMLM 313
+++ W + + + + Q V + AM V +G+SN+ +G ML+
Sbjct 831 VIYSIFYTWNVKKSMINIRTQMFPLVFPRWDIFGAMTSVFMFTYLYAEGKSNYFKGSMLI 890
Query 314 AAYLIISVVFWFD 326
Y+II V F+F
Sbjct 891 LLYIIIVVGFYFQ 903
> dre:560283 cax1, MGC136271, si:dkey-180p18.2, wu:fc25f10, zgc:136271;
cation/H+ exchanger protein 1
Length=764
Score = 103 bits (257), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 93/369 (25%), Positives = 162/369 (43%), Gaps = 54/369 (14%)
Query 12 ALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQAL----RIG---LLEVV 64
++IPLS +G +S + G ++NATFG+ EM + AL R G E+V
Sbjct 397 SIIPLSYYIGMGIASISAQSNFAVGAVVNATFGSITEMAFYITALLQGQRAGSQCYAELV 456
Query 65 KGTLLGSILSNLLLVLGMSFFAGGLHHYLQKFNEKGATCSVTLLLLSCMGIVIPTVAAVD 124
K L G++L +L + G+ GG H Q+FN + A S LL +S G+ PT+ +
Sbjct 457 KSALTGTLLGCILFIPGLCMIIGGFKHSEQRFNSRSAGVSSALLFISVGGVFAPTLFSKA 516
Query 125 NGQ--------------------HGTYNI------LMISRITALLIGVT------YCLFL 152
G + Y++ L +S I L+ V+ Y + L
Sbjct 517 YGNFICEGCNNPPGNISKPFICNNCHYDLSENNRPLFLSHIEPLVYTVSVLLPAAYLVGL 576
Query 153 FFQLYTHIGLFK-----------DDDEDAEQWPMMSWEAATLMLFLVTSLVALHSELLVS 201
F L TH ++ D W S A ++L L T L+A ++L
Sbjct 577 IFTLKTHSHIYDIHISDGHCHVISDHHAVAHW---SRTRALVVLILATILMAACADLSTE 633
Query 202 SIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVDLAMGVAVGSSAQIALFV 261
I+ ++++ +S+ FIGV ++ +V E + V A++N + L++ V + Q+ +
Sbjct 634 HIKPIISNSSVSQYFIGVTIIAMVPELPEIVNGVQFALQNNISLSLEVGSCLAVQVCMLQ 693
Query 262 FPFTVLVAWVMGQPLTMAVQPLSAVVMLLSVLVAMAVVQDGESNWLEGVMLMAAYLIISV 321
P +L + L + SV++ + DG+S++ +G L+ YLI+
Sbjct 694 IPLLILFNAFYDVGFVLIFSDLHLWASIFSVILVNYIFMDGKSDYFQGTALVVVYLILLA 753
Query 322 VFWFDKPGP 330
+++F P P
Sbjct 754 LYFF-APAP 761
> xla:734450 hypothetical protein MGC115264
Length=750
Score = 96.7 bits (239), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 86/371 (23%), Positives = 160/371 (43%), Gaps = 47/371 (12%)
Query 6 FSFNFAALIPLSALLGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLE--- 62
F+ + +++PLS +G +S + G ++NATFG+ +E+ + AL G E
Sbjct 378 FTLSLVSIMPLSYYIGMAIASVSAQSNFAVGAVVNATFGSIIELTFYITALIKGAREGNR 437
Query 63 ----VVKGTLLGSILSNLLLVLGMSFFAGGLHHYLQKFNEKGATCSVTLLLLSCMGIVIP 118
VVK L G++L +L + G+ GG+ H Q+FN + A S LL +S G+ P
Sbjct 438 CYEEVVKSALTGTVLGCVLFIPGLCMIIGGIKHREQRFNSRSAGVSSALLFISVGGVFAP 497
Query 119 TVAA-----------------------VDNGQHG---TYNILMISRITALLIGVT----- 147
T+ + N Q G T + S + L+ V+
Sbjct 498 TLFSKVYGKMICGECMNFTQNTTEKYICSNCQTGIMETNSTAFYSEVKPLVYTVSILLPA 557
Query 148 -YCLFLFFQLYTHIGLFKDDDEDA-----EQWPMMSWEAATLMLFLV--TSLVALHSELL 199
Y + L F L TH ++ D ++ W M+ L+ T L++ ++L
Sbjct 558 AYIIGLLFTLKTHSHIYDIHVSDCHVPGHHHSAVVHWSRWRAMIILLGSTFLMSACADLA 617
Query 200 VSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVDLAMGVAVGSSAQIAL 259
I ++ +S+ F+GV +L ++ E + + A+ N + L++ + + Q+ +
Sbjct 618 TEHISPILTGATVSQYFVGVTILAMIPELPEIVNGIQFALLNNLSLSIEIGSCIAVQVCM 677
Query 260 FVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSVLVAMAVVQDGESNWLEGVMLMAAYLII 319
P VL + T+ L + SV++ + DG+ ++ +G L+ Y I+
Sbjct 678 LQIPILVLFSVFYATDFTLIFSDLHLWASMFSVILMNYIFMDGKCDYFQGTALVFVYFIL 737
Query 320 SVVFWFDKPGP 330
+++F P P
Sbjct 738 LAMYFF-APSP 747
> eco:b1216 chaA, ECK1210, JW1207; calcium/sodium:proton antiporter;
K07300 Ca2+:H+ antiporter
Length=366
Score = 43.9 bits (102), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 70/255 (27%), Positives = 114/255 (44%), Gaps = 25/255 (9%)
Query 79 VLGMSFFAGGLHHYLQKFNEKG-ATCSVTLLLLSCMGIVIPTVAAVDNGQHGTYNILMIS 137
++G S GG Q N G + L L+ + +V P A+ T L+++
Sbjct 118 LVGFSLLLGGRKFATQYMNLFGIKQYLIALFPLAIIVLVFPM--ALPAANFSTGQALLVA 175
Query 138 RITALLIGVTYCLFLFFQLYTHIGLFKDDDEDA-------EQWPMMS---WEAATLMLFL 187
I+A + GV FL Q TH LF + ED P W A L++ L
Sbjct 176 LISAAMYGV----FLLIQTKTHQSLFVYEHEDDSDDDDPHHGKPSAHSSLWHAIWLIIHL 231
Query 188 VTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVDLAM 247
+ + +++ SS+E ++ +F G L+ ++ + E L A+ + N+V AM
Sbjct 232 IAVIAV--TKMNASSLETLLDSMNAPVAFTG-FLVALLILSPEGLGALKAVLNNQVQRAM 288
Query 248 GVAVGSS-AQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSVLVAMAVVQDGESNW 306
+ GS A I+L V P L+A++ G L A+ VVM+ S+++ G +N
Sbjct 289 NLFFGSVLATISLTV-PVVTLIAFMTGNELQFALGAPEMVVMVASLVLCHISFSTGRTNV 347
Query 307 LEG---VMLMAAYLI 318
L G + L AAYL+
Sbjct 348 LNGAAHLALFAAYLM 362
> dre:561209 solute carrier family 24 (sodium/potassium/calcium
exchanger), member 3-like
Length=639
Score = 42.0 bits (97), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 33/121 (27%), Positives = 58/121 (47%), Gaps = 5/121 (4%)
Query 198 LLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVDLAMGVAVGSSAQI 257
V S+E++ + LSE G + +A E T++ K DL +G VGS+
Sbjct 128 FFVPSLEKICENLHLSEDVAGATFMAAGSSAPELFTSLIGVFITKGDLGVGTIVGSAVFN 187
Query 258 ALFVFPFTVLVAWVMGQPLTMAVQPL--SAVVMLLSVLVAMAVVQDGESNWLEGVMLMAA 315
L + + A GQ +T+ PL ++ +LSVL + ++ D + W E ++L++
Sbjct 188 ILVIIGVCGIFA---GQTITLTWWPLFRDSIFYILSVLALILIIYDEKVIWWETIILISM 244
Query 316 Y 316
Y
Sbjct 245 Y 245
> hsa:57419 SLC24A3, NCKX3; solute carrier family 24 (sodium/potassium/calcium
exchanger), member 3; K13751 solute carrier
family 24 (sodium/potassium/calcium exchanger), member 3
Length=644
Score = 38.5 bits (88), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 35/144 (24%), Positives = 67/144 (46%), Gaps = 5/144 (3%)
Query 185 LFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVD 244
+++ +L + + V S+E++ LSE G + +A E T+V K D
Sbjct 118 IYMFYALAIVCDDFFVPSLEKICERLHLSEDVAGATFMAAGSSAPELFTSVIGVFITKGD 177
Query 245 LAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPL--SAVVMLLSVLVAMAVVQDG 302
+ +G VGS+ L + L A GQ + ++ L ++ LSV+ + + D
Sbjct 178 VGVGTIVGSAVFNILCIIGVCGLFA---GQVVALSSWCLLRDSIYYTLSVIALIVFIYDE 234
Query 303 ESNWLEGVMLMAAYLIISVVFWFD 326
+ +W E ++L+ YLI V+ ++
Sbjct 235 KVSWWESLVLVLMYLIYIVIMKYN 258
> mmu:94249 Slc24a3, NCKX3; solute carrier family 24 (sodium/potassium/calcium
exchanger), member 3; K13751 solute carrier
family 24 (sodium/potassium/calcium exchanger), member 3
Length=595
Score = 38.5 bits (88), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 35/144 (24%), Positives = 67/144 (46%), Gaps = 5/144 (3%)
Query 185 LFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVD 244
+++ +L + + V S+E++ LSE G + +A E T+V K D
Sbjct 67 MYMFYALAIVCDDFFVPSLEKICERLHLSEDVAGATFMAAGSSAPELFTSVIGVFITKGD 126
Query 245 LAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPL--SAVVMLLSVLVAMAVVQDG 302
+ +G VGS+ L + L A GQ + ++ L ++ LSV+ + + D
Sbjct 127 VGVGTIVGSAVFNILCIIGVCGLFA---GQVVALSSWCLLRDSIYYTLSVVALIVFIYDE 183
Query 303 ESNWLEGVMLMAAYLIISVVFWFD 326
+ +W E ++L+ YLI V+ ++
Sbjct 184 KVSWWESLVLVLMYLIYIVIMKYN 207
> dre:570312 slc24a5; solute carrier family 24, member 5; K13753
solute carrier family 24 (sodium/potassium/calcium exchanger),
member 5
Length=513
Score = 38.1 bits (87), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 33/146 (22%), Positives = 65/146 (44%), Gaps = 5/146 (3%)
Query 183 LMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNK 242
++ +++ S+ + E + S+E + GLS+ G + +A E +TA K
Sbjct 87 IIFYMLLSVSIVCDEYFLPSLEVISERLGLSQDVAGATFMAAGSSAPELVTAFLGVFVTK 146
Query 243 VDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPL--SAVVMLLSVLVAMAVVQ 300
D+ + +GS+ L + L++ +G+ ++ PL V +SV +A++
Sbjct 147 GDIGVSTIMGSAVYNLLCICAACGLLSSAVGR---LSCWPLFRDCVAYAISVAAVIAIIS 203
Query 301 DGESNWLEGVMLMAAYLIISVVFWFD 326
D W +G L+ Y + V FD
Sbjct 204 DNRVYWYDGACLLLVYGVYVAVLCFD 229
> mmu:214111 Slc24a1, MGC27617; solute carrier family 24 (sodium/potassium/calcium
exchanger), member 1; K13749 solute carrier
family 24 (sodium/potassium/calcium exchanger), member
1
Length=1130
Score = 36.6 bits (83), Expect = 0.13, Method: Composition-based stats.
Identities = 30/104 (28%), Positives = 54/104 (51%), Gaps = 10/104 (9%)
Query 185 LFLVTSL-----VALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAM 239
F++T L +A+ S L+V +V G+SE +G+ +L + + +T+V VA
Sbjct 967 FFVITFLGSIIWIAMFSYLMVWWAHQVGETIGISEEIMGLTILAAGTSIPDLITSVIVAR 1026
Query 240 KNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPL 283
K D+A+ +VGS+ +F + V W++ L A+QP+
Sbjct 1027 KGLGDMAVSSSVGSN----IFDITVGLPVPWLLFS-LINALQPV 1065
Score = 32.3 bits (72), Expect = 2.4, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 34/62 (54%), Gaps = 1/62 (1%)
Query 20 LGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVVKGTLLGSILSNLLLV 79
LG T +L + + ++ G A G+A E+ S+ + I V GT++GS + N+L V
Sbjct 448 LGVITHKLQI-SEDVAGATFMAAGGSAPELFTSLIGVFISHSNVGIGTIVGSAVFNILFV 506
Query 80 LG 81
+G
Sbjct 507 IG 508
> mmu:238384 Slc24a4, A930002M03Rik, AW492432, NCKX4; solute carrier
family 24 (sodium/potassium/calcium exchanger), member
4; K13752 solute carrier family 24 (sodium/potassium/calcium
exchanger), member 4
Length=605
Score = 35.8 bits (81), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 36/135 (26%), Positives = 59/135 (43%), Gaps = 5/135 (3%)
Query 185 LFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVD 244
L++ +L + + V S+E++ LSE G + + E +V D
Sbjct 91 LYMFYALAIVCDDFFVPSLEKICEKLHLSEDVAGATFMAAGSSTPELFASVIGVFITHGD 150
Query 245 LAMGVAVGSSAQIALFVFPFTVLVAWVMGQ--PLTMAVQPLSAVVMLLSVLVAMAVVQDG 302
+ +G VGS+ L + L A GQ LT +V LSV+V +A + D
Sbjct 151 VGVGTIVGSAVFNILCIIGVCGLFA---GQVVRLTWWAVCRDSVYYTLSVIVLIAFIYDE 207
Query 303 ESNWLEGVMLMAAYL 317
E W EG++L+ Y+
Sbjct 208 EIVWWEGLVLIILYV 222
> dre:557196 slc24a3, si:dkey-187a12.3; solute carrier family
24 (sodium/potassium/calcium exchanger), member 3; K13751 solute
carrier family 24 (sodium/potassium/calcium exchanger),
member 3
Length=559
Score = 34.3 bits (77), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 35/148 (23%), Positives = 64/148 (43%), Gaps = 13/148 (8%)
Query 185 LFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVD 244
+++ +L + + V S+E++ LSE G + +A E T+V K D
Sbjct 32 MYMFYALALVCDDYFVPSLEKICERLHLSEDVAGATFMAAGSSAPELFTSVIGVFITKGD 91
Query 245 LAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSVLVAMAV------ 298
+ +G VGS+ VF ++ V G AV+ +M S +AV
Sbjct 92 VGVGTIVGSA------VFNILCIIG-VCGIFTAQAVRLSCWTLMRDSTYYTIAVATLIVF 144
Query 299 VQDGESNWLEGVMLMAAYLIISVVFWFD 326
+ D + W E ++L+ YL+ ++ F+
Sbjct 145 IYDEKVTWWESLILIVLYLVYILIMKFN 172
> hsa:283652 SLC24A5, JSX, NCKX5, SHEP4; solute carrier family
24, member 5; K13753 solute carrier family 24 (sodium/potassium/calcium
exchanger), member 5
Length=500
Score = 33.9 bits (76), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 33/144 (22%), Positives = 62/144 (43%), Gaps = 7/144 (4%)
Query 185 LFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVD 244
+F+ S+V E + S+E + GLS+ G + +A E +TA K D
Sbjct 79 MFMAISIVC--DEYFLPSLEIISESLGLSQDVAGTTFMAAGSSAPELVTAFLGVFITKGD 136
Query 245 LAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPL--SAVVMLLSVLVAMAVVQDG 302
+ + +GS+ L + L++ + T++ PL +S + ++ D
Sbjct 137 IGISTILGSAIYNLLGICAACGLLSNTVS---TLSCWPLFRDCAAYTISAAAVLGIIYDN 193
Query 303 ESNWLEGVMLMAAYLIISVVFWFD 326
+ W EG +L+ Y + +V FD
Sbjct 194 QVYWYEGALLLLIYGLYVLVLCFD 217
> dre:436697 slc48a1a, zgc:92662; solute carrier family 48 (heme
transporter), member 1a
Length=144
Score = 32.7 bits (73), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 33/62 (53%), Gaps = 5/62 (8%)
Query 264 FTVLVAW--VMGQPLTMAVQPLSAVVMLLSVLVAMAVVQDGESNWLEGVMLMAAYLIISV 321
F+ + W V QP T A+ LS V+ L +++ + +QD WL+G+ ++ +S
Sbjct 21 FSAFIVWNVVYKQPWTAAMGGLSGVLALWALVTHIMYIQDYWRTWLKGLKF---FMFVSS 77
Query 322 VF 323
VF
Sbjct 78 VF 79
> cel:C55C3.1 hypothetical protein
Length=606
Score = 32.7 bits (73), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 29/92 (31%), Positives = 43/92 (46%), Gaps = 12/92 (13%)
Query 39 LNATFGNAVEMIMSVQALRIGLLEVVKGTLLGSILSNLLLVLGMS----FFAGGLHHYLQ 94
LN+T N + M Q + IGLL G L GS+ N+++ L ++ A GL +L+
Sbjct 108 LNSTAPNKISWTM--QNMDIGLL----GDLSGSV--NIVVPLNLTGQVEILAQGLTFHLE 159
Query 95 KFNEKGATCSVTLLLLSCMGIVIPTVAAVDNG 126
EKG S + LSC+ + NG
Sbjct 160 SSIEKGKNGSAKVTSLSCLATIRDVTVTNHNG 191
> dre:558299 slc24a1, si:ch211-117i10.3; solute carrier family
24 (sodium/potassium/calcium exchanger), member 1; K13749 solute
carrier family 24 (sodium/potassium/calcium exchanger),
member 1
Length=724
Score = 32.3 bits (72), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 37/63 (58%), Gaps = 1/63 (1%)
Query 20 LGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVVKGTLLGSILSNLLLV 79
LG T++L++ + ++ G A G+A E+ S+ + I V GT++GS + N+L V
Sbjct 196 LGVITDKLAI-SDDVAGATFMAAGGSAPELFTSLIGVFISHSNVGIGTIVGSAVFNILFV 254
Query 80 LGM 82
+GM
Sbjct 255 IGM 257
Score = 31.2 bits (69), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 50/98 (51%), Gaps = 9/98 (9%)
Query 180 AATLMLFLVTSL-----VALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTA 234
A+ F++T L +A+ S L+V +V G+SE +G+ +L + + +T+
Sbjct 556 PASKKFFVITFLGSIVWIAIFSYLMVWWAHQVGETIGISEEIMGLTILAAGTSIPDLITS 615
Query 235 VTVAMKNKVDLAMGVAVGSSAQIALFVFPFTVLVAWVM 272
V VA K D+A+ +VGS+ +F + V W+M
Sbjct 616 VIVARKGLGDMAVSSSVGSN----IFDITMGLPVPWLM 649
> dre:571043 NCKX2.3-like
Length=276
Score = 32.0 bits (71), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 36/63 (57%), Gaps = 1/63 (1%)
Query 20 LGAFTEELSLHTGEITGGLLNATFGNAVEMIMSVQALRIGLLEVVKGTLLGSILSNLLLV 79
L TE+L++ + ++ G A G+A E+ SV + I V GT++GS + N+L V
Sbjct 130 LTVITEKLTI-SDDVAGATFMAAGGSAPELFTSVIGVFISHSNVGIGTIVGSAVFNILFV 188
Query 80 LGM 82
+GM
Sbjct 189 IGM 191
> hsa:123041 SLC24A4, FLJ38852, NCKX4, SHEP6, SLC24A2; solute
carrier family 24 (sodium/potassium/calcium exchanger), member
4; K13752 solute carrier family 24 (sodium/potassium/calcium
exchanger), member 4
Length=622
Score = 31.2 bits (69), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 33/135 (24%), Positives = 58/135 (42%), Gaps = 5/135 (3%)
Query 185 LFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKVD 244
L++ +L + + V S+E++ LSE G + + E +V D
Sbjct 108 LYMFYALAIVCDDFFVPSLEKICERLHLSEDVAGATFMAAGSSTPELFASVIGVFITHGD 167
Query 245 LAMGVAVGSSAQIALFVFPFTVLVAWVMGQ--PLTMAVQPLSAVVMLLSVLVAMAVVQDG 302
+ +G VGS+ L + L A GQ LT +V +SV+V + + D
Sbjct 168 VGVGTIVGSAVFNILCIIGVCGLFA---GQVVRLTWWAVCRDSVYYTISVIVLIVFIYDE 224
Query 303 ESNWLEGVMLMAAYL 317
+ W EG++L+ Y+
Sbjct 225 QIVWWEGLVLIILYV 239
> dre:792763 slc48a1b, MGC174054, MGC63994, fj61c11, hrg1, wu:fj61c11,
zgc:63994; solute carrier family 48 (heme transporter),
member 1b
Length=144
Score = 30.8 bits (68), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 33/62 (53%), Gaps = 3/62 (4%)
Query 264 FTVLVAW--VMGQPLTMAVQPLSAVVMLLSVLVAMAVVQDGESNWLEGV-MLMAAYLIIS 320
F+ + W QP T A+ LS V+ L +++ + +QD WL+G+ MA +I S
Sbjct 21 FSAFLVWNIAYKQPWTAAMGGLSGVLALWALVTHIMYIQDYWRTWLKGLKFFMAMGVIFS 80
Query 321 VV 322
V+
Sbjct 81 VL 82
> xla:494820 slc48a1-a, HRG-1A, hHRG-1, hrg1, hrg1-a, slc48a1;
solute carrier family 48 (heme transporter), member 1
Length=145
Score = 30.8 bits (68), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query 274 QPLTMAVQPLSAVVMLLSVLVAMAVVQDGESNWLEGVMLMAAYLIISVVF 323
QP T A+ LS V+ L +++ + VQD WL+G+ +L I V+F
Sbjct 34 QPWTAAMGGLSGVLALWALITHIMYVQDFWRTWLKGLRF---FLCIGVLF 80
> ath:ArthCp030 rbcL; RuBisCO large subunit; K01601 ribulose-bisphosphate
carboxylase large chain [EC:4.1.1.39]
Length=479
Score = 30.8 bits (68), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 21/49 (42%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 31 TGEITGGLLNATFGNAVEMI-MSVQALRIGLLEVVKGTLLGSILSNLLL 78
TGEI G LNAT G EMI +V A +G+ V+ L G +N L
Sbjct 232 TGEIKGHYLNATAGTCEEMIKRAVFARELGVPIVMHDYLTGGFTANTSL 280
> xla:735139 slc48a1-b, HRG-1B, MGC115395, hHRG-1, hrg1, hrg1-b;
solute carrier family 48 (heme transporter), member 1
Length=145
Score = 30.4 bits (67), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query 274 QPLTMAVQPLSAVVMLLSVLVAMAVVQDGESNWLEGVMLMAAYLIISVVF 323
QP T A+ LS V+ L +++ + VQD WL+G+ +L I V+F
Sbjct 34 QPWTAAMGGLSGVLALWALITHIMYVQDFWRTWLKGLRF---FLCIGVLF 80
> dre:550467 zgc:112072; K13752 solute carrier family 24 (sodium/potassium/calcium
exchanger), member 4
Length=569
Score = 30.4 bits (67), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 36/148 (24%), Positives = 64/148 (43%), Gaps = 7/148 (4%)
Query 181 ATLMLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMK 240
A L +FL ++V + V+S+E++ LSE G + +A E ++
Sbjct 90 AALYMFLALAIVC--DDYFVTSLEKICEKLHLSEDVAGATFMAAGSSAPELFASIIGVFI 147
Query 241 NKVDLAMGVAVGSSAQIALFVFPFTVLVAWVMGQP--LTMAVQPLSAVVMLLSVLVAMAV 298
D+ +G VGS+ L + + A GQ LT + ++SV+ +
Sbjct 148 THGDVGVGTIVGSAVFNILCIIGVCGIFA---GQVVFLTKYAVFRDSSYYIISVIALIVF 204
Query 299 VQDGESNWLEGVMLMAAYLIISVVFWFD 326
+ D E W E ++L+ YL +V F+
Sbjct 205 IYDDEILWWESLVLIVMYLGYIIVMKFN 232
> hsa:25769 SLC24A2, FLJ45149, NCKX2; solute carrier family 24
(sodium/potassium/calcium exchanger), member 2; K13750 solute
carrier family 24 (sodium/potassium/calcium exchanger), member
2
Length=644
Score = 30.4 bits (67), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 34/145 (23%), Positives = 61/145 (42%), Gaps = 5/145 (3%)
Query 184 MLFLVTSLVALHSELLVSSIEEVVADYGLSESFIGVILLPIVGNAAEHLTAVTVAMKNKV 243
M+++ +L + E V S+ + G+S+ G + G+A E T++
Sbjct 142 MIYMFIALAIVCDEFFVPSLTVITEKLGISDDVAGATFMAAGGSAPELFTSLIGVFIAHS 201
Query 244 DLAMGVAVGSSAQIALFVFPFTVLVAWVMGQPLTMAVQPLSAVVMLLSVLVAMAVV--QD 301
++ +G VGS+ LFV L + + L + PL V V + M ++ D
Sbjct 202 NVGIGTIVGSAVFNILFVIGMCALFS---REILNLTWWPLFRDVSFYIVDLIMLIIFFLD 258
Query 302 GESNWLEGVMLMAAYLIISVVFWFD 326
W E ++L+ AY V F+
Sbjct 259 NVIMWWESLLLLTAYFCYVVFMKFN 283
Lambda K H
0.325 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 14410718180
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40