bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2186_orf1
Length=133
Score E
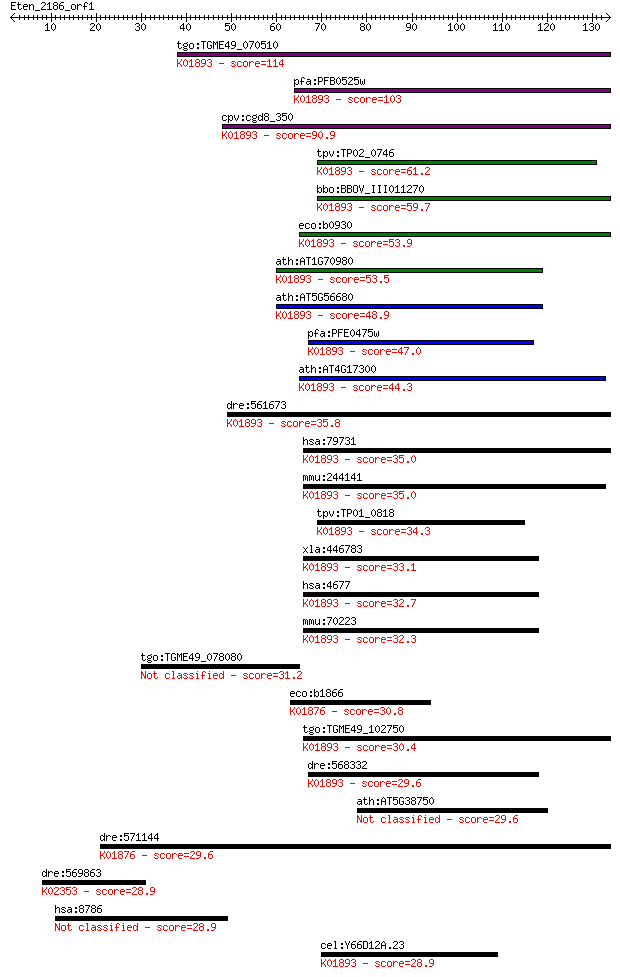
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_070510 asparaginyl-tRNA synthetase, putative (EC:6.... 114 9e-26
pfa:PFB0525w asparagine-tRNA ligase, putative; K01893 asparagi... 103 1e-22
cpv:cgd8_350 asparaginyl-tRNA synthetase (NOB+tRNA synthase) ;... 90.9 9e-19
tpv:TP02_0746 asparaginyl-tRNA synthetase; K01893 asparaginyl-... 61.2 9e-10
bbo:BBOV_III011270 17.m07970; asparaginyl-tRNA synthetase fami... 59.7 3e-09
eco:b0930 asnS, ECK0921, JW0913, lcs, tss; asparaginyl tRNA sy... 53.9 1e-07
ath:AT1G70980 SYNC3; SYNC3; ATP binding / aminoacyl-tRNA ligas... 53.5 2e-07
ath:AT5G56680 SYNC1; SYNC1; ATP binding / aminoacyl-tRNA ligas... 48.9 4e-06
pfa:PFE0475w asparagine-tRNA ligase, putative (EC:6.1.1.22); K... 47.0 2e-05
ath:AT4G17300 NS1; NS1; asparagine-tRNA ligase (EC:6.1.1.22); ... 44.3 1e-04
dre:561673 MGC153218; zgc:153218 (EC:6.1.1.22); K01893 asparag... 35.8 0.036
hsa:79731 NARS2, FLJ23441, SLM5; asparaginyl-tRNA synthetase 2... 35.0 0.068
mmu:244141 Nars2, AI875199, MGC41336; asparaginyl-tRNA synthet... 35.0 0.069
tpv:TP01_0818 asparaginyl-tRNA synthetase; K01893 asparaginyl-... 34.3 0.10
xla:446783 nars, MGC80438; asparaginyl-tRNA synthetase (EC:6.1... 33.1 0.23
hsa:4677 NARS, ASNRS, NARS1; asparaginyl-tRNA synthetase (EC:6... 32.7 0.29
mmu:70223 Nars, 3010001M15Rik, AA960128, ASNRS, C78150; aspara... 32.3 0.42
tgo:TGME49_078080 hypothetical protein 31.2 0.84
eco:b1866 aspS, ECK1867, JW1855, tls; aspartyl-tRNA synthetase... 30.8 1.2
tgo:TGME49_102750 OB-fold nucleic acid binding domain-containi... 30.4 1.4
dre:568332 nars, fb57a10, si:dkey-274m14.2, wu:fb57a10; aspara... 29.6 2.4
ath:AT5G38750 asparaginyl-tRNA synthetase family 29.6 2.5
dre:571144 dars2, si:dkey-21n10.1; aspartyl-tRNA synthetase 2,... 29.6 2.8
dre:569863 dvl1a, si:dkey-223i19.4; dishevelled, dsh homolog 1... 28.9 4.5
hsa:8786 RGS11, RS11; regulator of G-protein signaling 11 28.9
cel:Y66D12A.23 hypothetical protein; K01893 asparaginyl-tRNA s... 28.9 4.9
> tgo:TGME49_070510 asparaginyl-tRNA synthetase, putative (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=676
Score = 114 bits (285), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 61/101 (60%), Positives = 76/101 (75%), Gaps = 5/101 (4%)
Query 38 QPIAVYGPGGGRMRLAALLASCAAA--TFIGRQ--VTVCGWSRSIRKQGGGSLCFVVLTD 93
Q I V+G G R+RLAALL A+ F+ R+ VTVCGWSRS+RKQGGGSLCFVVL+D
Sbjct 159 QAIPVFGCGSTRVRLAALLDGGEASVEKFVNRETPVTVCGWSRSVRKQGGGSLCFVVLSD 218
Query 94 GSCAANLQVVVNKTI-PKFDELIKCGVGCSFKFCGRVVESP 133
GS ++NLQVVV I +F +L+KCG GCSF+F G +V+SP
Sbjct 219 GSTSSNLQVVVEAGIGGEFPQLLKCGAGCSFRFTGDIVKSP 259
> pfa:PFB0525w asparagine-tRNA ligase, putative; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=610
Score = 103 bits (258), Expect = 1e-22, Method: Composition-based stats.
Identities = 41/70 (58%), Positives = 55/70 (78%), Gaps = 0/70 (0%)
Query 64 FIGRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCGVGCSF 123
+IG+ +TVCGWS++IRKQGGG CFV L DGSC NLQ+VVN+ I +++L+KCG GC F
Sbjct 86 YIGKIITVCGWSKAIRKQGGGRFCFVNLNDGSCHLNLQIVVNQCIENYEKLLKCGAGCCF 145
Query 124 KFCGRVVESP 133
+F G ++ SP
Sbjct 146 RFTGELIISP 155
> cpv:cgd8_350 asparaginyl-tRNA synthetase (NOB+tRNA synthase)
; K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=499
Score = 90.9 bits (224), Expect = 9e-19, Method: Composition-based stats.
Identities = 41/87 (47%), Positives = 61/87 (70%), Gaps = 1/87 (1%)
Query 48 GRMRLAALL-ASCAAATFIGRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNK 106
GR+R+A++L ++IG++VTV GW+R++RKQ +L F+ L DGS ++NLQ VV K
Sbjct 16 GRLRIASILDGEDGGVSYIGKKVTVGGWARTVRKQCSDTLLFISLNDGSTSSNLQCVVEK 75
Query 107 TIPKFDELIKCGVGCSFKFCGRVVESP 133
T+ F+E +K GCSFK G +V+SP
Sbjct 76 TVKGFEEGLKATAGCSFKITGTIVKSP 102
> tpv:TP02_0746 asparaginyl-tRNA synthetase; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=710
Score = 61.2 bits (147), Expect = 9e-10, Method: Composition-based stats.
Identities = 28/62 (45%), Positives = 39/62 (62%), Gaps = 0/62 (0%)
Query 69 VTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCGVGCSFKFCGR 128
V V GW +S+R Q GG L FV++ DGSC +NLQ+VV+ + E +KC G S + G
Sbjct 143 VRVFGWCKSMRSQDGGRLLFVIVNDGSCKSNLQIVVHNDSKGYKEALKCKSGTSIQANGF 202
Query 129 VV 130
+V
Sbjct 203 LV 204
> bbo:BBOV_III011270 17.m07970; asparaginyl-tRNA synthetase family
protein; K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=605
Score = 59.7 bits (143), Expect = 3e-09, Method: Composition-based stats.
Identities = 27/65 (41%), Positives = 39/65 (60%), Gaps = 0/65 (0%)
Query 69 VTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCGVGCSFKFCGR 128
++V GW +++R Q G L FV+L DGSC A+LQVVV++ + KC G + G
Sbjct 95 ISVAGWCKTVRMQNAGKLGFVILNDGSCKADLQVVVHEGAIGHQAVDKCTSGTTIAIRGT 154
Query 129 VVESP 133
+VE P
Sbjct 155 LVERP 159
> eco:b0930 asnS, ECK0921, JW0913, lcs, tss; asparaginyl tRNA
synthetase (EC:6.1.1.22); K01893 asparaginyl-tRNA synthetase
[EC:6.1.1.22]
Length=466
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 42/70 (60%), Gaps = 2/70 (2%)
Query 65 IGRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDE-LIKCGVGCSF 123
+ +VTV GW R+ R G + F+ + DGSC +Q V+N ++P ++E +++ GCS
Sbjct 16 VDSEVTVRGWVRTRRDSKAG-ISFLAVYDGSCFDPVQAVINNSLPNYNEDVLRLTTGCSV 74
Query 124 KFCGRVVESP 133
G+VV SP
Sbjct 75 IVTGKVVASP 84
> ath:AT1G70980 SYNC3; SYNC3; ATP binding / aminoacyl-tRNA ligase/
asparagine-tRNA ligase/ aspartate-tRNA ligase/ nucleic
acid binding / nucleotide binding (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=571
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 60 AAATFIGRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCG 118
A G++V + GW ++ R+QG G+ F+ + DGSC ANLQV+V+ ++ L+ G
Sbjct 41 GGAKLAGQKVRIGGWVKTGRQQGKGTFAFLEVNDGSCPANLQVMVDSSLYDLSRLVATG 99
> ath:AT5G56680 SYNC1; SYNC1; ATP binding / aminoacyl-tRNA ligase/
asparagine-tRNA ligase/ aspartate-tRNA ligase/ nucleic
acid binding / nucleotide binding (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=572
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 60 AAATFIGRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCG 118
A G+ V + GW +S R QG + F+ + DGSC ANLQV+V+ ++ L+ G
Sbjct 44 GGAGLAGQTVRIGGWVKSGRDQGKRTFSFLAVNDGSCPANLQVMVDPSLYDVSNLVATG 102
> pfa:PFE0475w asparagine-tRNA ligase, putative (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=722
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 30/50 (60%), Gaps = 0/50 (0%)
Query 67 RQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIK 116
+ +CGW +S+R G FV + DGS NLQV+++ IP ++ ++K
Sbjct 120 KNFKICGWIKSVRTVGKSYFVFVDINDGSYYKNLQVIIDDKIPDYEYILK 169
> ath:AT4G17300 NS1; NS1; asparagine-tRNA ligase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=567
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 38/70 (54%), Gaps = 4/70 (5%)
Query 65 IGRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCGV--GCS 122
+G+ + + GW R++R Q S+ F+ + DGSC +NLQ V+ +D++ + G S
Sbjct 108 VGQSLNIMGWVRTLRSQS--SVTFIEINDGSCLSNLQCVMTSDAEGYDQVESGSILTGAS 165
Query 123 FKFCGRVVES 132
G +V S
Sbjct 166 VSVQGTIVAS 175
> dre:561673 MGC153218; zgc:153218 (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=485
Score = 35.8 bits (81), Expect = 0.036, Method: Composition-based stats.
Identities = 28/85 (32%), Positives = 39/85 (45%), Gaps = 10/85 (11%)
Query 49 RMRLAALLASCAAATFIGRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTI 108
+MR++ LA C G V + GW RS+R Q F+ L DGS LQVV + +
Sbjct 35 KMRVSDALA-CKET---GSDVRIQGWVRSVRPQKDN--LFLHLNDGSSLQPLQVVASSQL 88
Query 109 PKFDELIKCGVGCSFKFCGRVVESP 133
D GC+ G + +SP
Sbjct 89 NTRD----LTFGCAVDITGTLEKSP 109
> hsa:79731 NARS2, FLJ23441, SLM5; asparaginyl-tRNA synthetase
2, mitochondrial (putative) (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=477
Score = 35.0 bits (79), Expect = 0.068, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 36/68 (52%), Gaps = 6/68 (8%)
Query 66 GRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCGVGCSFKF 125
G ++ + GW RS+R Q + F+ + DGS +LQVV + + + G S +
Sbjct 41 GERIKIQGWIRSVRSQ--KEVLFLHVNDGSSLESLQVVADSGLDSRE----LNFGSSVEV 94
Query 126 CGRVVESP 133
G++++SP
Sbjct 95 QGQLIKSP 102
> mmu:244141 Nars2, AI875199, MGC41336; asparaginyl-tRNA synthetase
2 (mitochondrial)(putative) (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=477
Score = 35.0 bits (79), Expect = 0.069, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 37/67 (55%), Gaps = 6/67 (8%)
Query 66 GRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCGVGCSFKF 125
G VTV GW RS+R Q + F+ + DGS +LQ+V + + FD + G S +
Sbjct 41 GECVTVQGWIRSVRSQ--KEVLFLHVNDGSSLESLQIVADSS---FDSR-ELTFGSSVQV 94
Query 126 CGRVVES 132
G++V+S
Sbjct 95 QGQLVKS 101
> tpv:TP01_0818 asparaginyl-tRNA synthetase; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=558
Score = 34.3 bits (77), Expect = 0.10, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Query 69 VTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDEL 114
V G+ + IR+ +L F+ + DGS ANLQ+VVN+ F EL
Sbjct 118 VNTRGFVQEIRRINSQNLYFIDINDGSTLANLQIVVNQA-NNFKEL 162
> xla:446783 nars, MGC80438; asparaginyl-tRNA synthetase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=559
Score = 33.1 bits (74), Expect = 0.23, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 31/52 (59%), Gaps = 9/52 (17%)
Query 66 GRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKC 117
G++V V GW +R+Q G +L F+VL DG+ LQ V++ D+L +C
Sbjct 136 GQRVKVFGWVHRLRRQ-GKNLMFIVLRDGT--GYLQCVLS------DQLCQC 178
> hsa:4677 NARS, ASNRS, NARS1; asparaginyl-tRNA synthetase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=548
Score = 32.7 bits (73), Expect = 0.29, Method: Composition-based stats.
Identities = 21/52 (40%), Positives = 30/52 (57%), Gaps = 9/52 (17%)
Query 66 GRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKC 117
G++V V GW +R+Q G +L F+VL DG+ LQ V+ DEL +C
Sbjct 125 GQRVKVFGWVHRLRRQ-GKNLMFLVLRDGT--GYLQCVLA------DELCQC 167
> mmu:70223 Nars, 3010001M15Rik, AA960128, ASNRS, C78150; asparaginyl-tRNA
synthetase (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=559
Score = 32.3 bits (72), Expect = 0.42, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 31/52 (59%), Gaps = 9/52 (17%)
Query 66 GRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKC 117
G++V V GW +R+Q G +L F+VL DG+ LQ V++ D+L +C
Sbjct 136 GQRVKVFGWVHRLRRQ-GKNLMFLVLRDGT--GYLQCVLS------DDLCQC 178
> tgo:TGME49_078080 hypothetical protein
Length=499
Score = 31.2 bits (69), Expect = 0.84, Method: Composition-based stats.
Identities = 16/35 (45%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 30 ETAALSYVQPIAVYGPGGGRMRLAALLASCAAATF 64
E+A L+ V A +G GGR + A + SCAAA F
Sbjct 462 ESAHLTIVIHSAAWGLAGGRAAIPAFVLSCAAALF 496
> eco:b1866 aspS, ECK1867, JW1855, tls; aspartyl-tRNA synthetase
(EC:6.1.1.12); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=590
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 21/31 (67%), Gaps = 2/31 (6%)
Query 63 TFIGRQVTVCGWSRSIRKQGGGSLCFVVLTD 93
+ +G+QVT+CGW R++ GSL F+ + D
Sbjct 12 SHVGQQVTLCGWVN--RRRDLGSLIFIDMRD 40
> tgo:TGME49_102750 OB-fold nucleic acid binding domain-containing
protein (EC:6.1.1.22); K01893 asparaginyl-tRNA synthetase
[EC:6.1.1.22]
Length=309
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 34/68 (50%), Gaps = 3/68 (4%)
Query 66 GRQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCGVGCSFKF 125
+V + GW +R+Q G SL F +L DG+ LQV++ + + + + C+ +
Sbjct 111 NERVCIKGWIHRMRRQ-GKSLMFFILRDGT--GFLQVLLMDKLCQTYDALTVNTECTVEI 167
Query 126 CGRVVESP 133
G + E P
Sbjct 168 YGAIKEVP 175
> dre:568332 nars, fb57a10, si:dkey-274m14.2, wu:fb57a10; asparaginyl-tRNA
synthetase; K01893 asparaginyl-tRNA synthetase
[EC:6.1.1.22]
Length=556
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 29/51 (56%), Gaps = 9/51 (17%)
Query 67 RQVTVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKC 117
++V V GW +R+Q G +L F+VL DG+ LQ V+ D+L +C
Sbjct 134 QRVKVFGWVHRLRRQ-GKNLLFIVLRDGT--GFLQCVLT------DKLCQC 175
> ath:AT5G38750 asparaginyl-tRNA synthetase family
Length=135
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 78 IRKQGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDELIKCGV 119
I ++ G S F+ + GSCAANLQV + E+I+ V
Sbjct 31 ISRELGKSFTFLEVNYGSCAANLQVKIPLQGKATKEMIELNV 72
> dre:571144 dars2, si:dkey-21n10.1; aspartyl-tRNA synthetase
2, mitochondrial; K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=660
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 25/114 (21%), Positives = 48/114 (42%), Gaps = 6/114 (5%)
Query 21 SPPALPPVSETAALSYVQPIAVYGPGGGRMRLAALLASCAAATFIGRQVTVCGWSRSIRK 80
S P + P A S + I+ + G + ++ I ++VT+CGW + +R+
Sbjct 39 SRPLISPNLLECAYSISRSISTHTSGQSSFSQRSHTCGELCSSHIDKEVTLCGWVQYLRQ 98
Query 81 QGGGSLCFVVLTDGSCAANLQVVVNKTIPKFDE-LIKCGVGCSFKFCGRVVESP 133
FV+L D S + + +++ + + L+ V GRV+ P
Sbjct 99 D-----LFVILRDFSGLVQILIPQDESKSELKKSLLALTVESVIMVKGRVIRRP 147
> dre:569863 dvl1a, si:dkey-223i19.4; dishevelled, dsh homolog
1a (Drosophila); K02353 dishevelled
Length=585
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 8 SHLFTMTMAGSAPSPPALPPVSE 30
+HL T AG++ SPP PP+ E
Sbjct 533 AHLAPKTAAGNSNSPPGAPPIRE 555
> hsa:8786 RGS11, RS11; regulator of G-protein signaling 11
Length=446
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 16/38 (42%), Positives = 19/38 (50%), Gaps = 8/38 (21%)
Query 11 FTMTMAGSAPSPPALPPVSETAALSYVQPIAVYGPGGG 48
FT S+PSP LP + V+P A GPGGG
Sbjct 413 FTWRPRHSSPSPALLP--------TPVEPTAACGPGGG 442
> cel:Y66D12A.23 hypothetical protein; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=448
Score = 28.9 bits (63), Expect = 4.9, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 2/39 (5%)
Query 70 TVCGWSRSIRKQGGGSLCFVVLTDGSCAANLQVVVNKTI 108
T+ GW++S++K G FV + G +Q+V NK I
Sbjct 14 TLDGWAKSVQKSGKN--VFVKIDKGRAGEPIQLVANKEI 50
Lambda K H
0.325 0.138 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2165519004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40