bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2211_orf3
Length=344
Score E
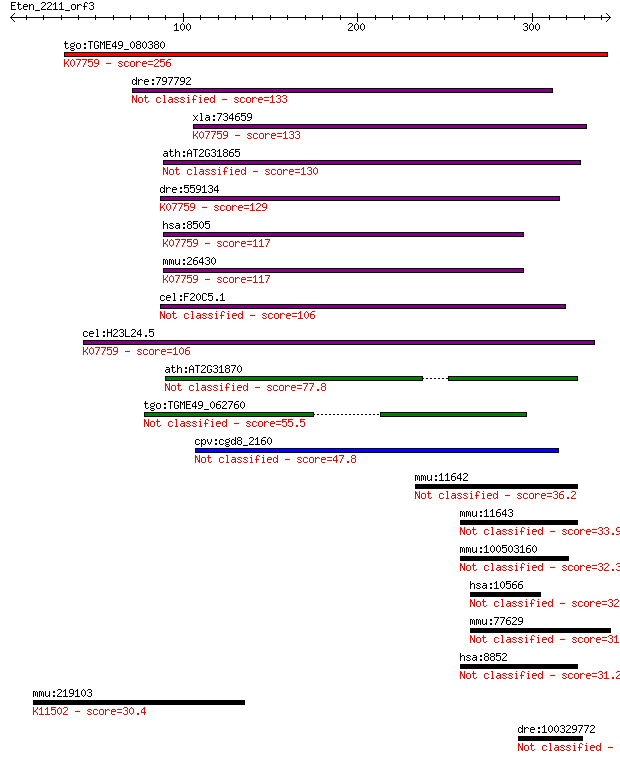
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143); K0... 256 1e-67
dre:797792 si:dkey-259k14.2 133 8e-31
xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase; ... 133 1e-30
ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family p... 130 9e-30
dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759... 129 1e-29
hsa:8505 PARG, FLJ54459, FLJ60257, FLJ60456, PARG99; poly (ADP... 117 6e-26
mmu:26430 Parg, AI413217; poly (ADP-ribose) glycohydrolase (EC... 117 7e-26
cel:F20C5.1 pme-3; Poly(ADP-ribose) Metabolism Enzyme family m... 106 1e-22
cel:H23L24.5 pme-4; Poly(ADP-ribose) Metabolism Enzyme family ... 106 1e-22
ath:AT2G31870 TEJ; TEJ (Sanskrit for 'bright'); poly(ADP-ribos... 77.8 6e-14
tgo:TGME49_062760 poly(ADP-ribose) glycohydrolase, putative 55.5 3e-07
cpv:cgd8_2160 shares a domain with poly(ADP) ribose glycohydro... 47.8 6e-05
mmu:11642 Akap3, Prka3; A kinase (PRKA) anchor protein 3 36.2
mmu:11643 Akap4, Fsc1, p82; A kinase (PRKA) anchor protein 4 33.9
mmu:100503160 a-kinase anchor protein 4-like 32.3 2.4
hsa:10566 AKAP3, AKAP110, CT82, FSP95, PRKA3, SOB1; A kinase (... 32.3 2.8
mmu:77629 Sphkap, 4930544G21Rik, A930009L15Rik, AI852220, mKIA... 31.6 5.2
hsa:8852 AKAP4, AKAP_82, AKAP-4, AKAP82, CT99, FSC1, HI, PRKA4... 31.2 5.3
mmu:219103 Cenpj, 4932437H03Rik, CPAP, Gm81, LAP, LIP1, MGC102... 30.4 9.3
dre:100329772 GTPase, IMAP family member 4-like 30.4 9.8
> tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=553
Score = 256 bits (653), Expect = 1e-67, Method: Compositional matrix adjust.
Identities = 126/317 (39%), Positives = 194/317 (61%), Gaps = 13/317 (4%)
Query 32 CRCTEVTFLGTKRHVAAADEVVSFYFQVPEETVFTSPEG-FEVVRPEIKLENVLQDKLPL 90
C C+ L +A DE++ FY Q + F +G + + ++++ + L
Sbjct 241 CICST---LEESVSIAPTDELIGFYRQSDKVADFVWSDGSVKGTSASVSIDDIAKSTKAL 297
Query 91 LPIQVHLNGDITKADGAVQVDFADRFLGGLTMHADHLAQEELIFTLKPELIVVMLFTQAL 150
+V GDIT +DG +QVDFAD++LGGL+M ++AQEELIF + PE++ VMLF+ +
Sbjct 298 GGFEVFEEGDITTSDGDLQVDFADKYLGGLSMFEGYIAQEELIFIIYPEMLCVMLFSDIM 357
Query 151 KADEAVVVKGAESFTLYRGYERTFELRAA-----EVPEGCHWSLHGLCPLDGLGRRAATV 205
K +EAVVVKG E F +GYE TF++ AA E+P H+S+ G+ PLD LGRR +
Sbjct 358 KPNEAVVVKGVERFVFSQGYEWTFQITAAAGDWTELPSNKHFSVQGVVPLDSLGRRNVAI 417
Query 206 AAIDALIVRNPNSQYQENKMKREVLKALLGFQGDPFEALLGEPKAPVATGKWGCVIFGGD 265
IDA+ PN QY + RE++KA +GF+GDP+E ++ + P+ATG WGC +F GD
Sbjct 418 VGIDAVQFHEPNKQYSPVMVNRELMKAAVGFKGDPYELIVSGSRQPIATGLWGCGVFNGD 477
Query 266 NQLKTVLQWIAASAAARPLQYMAFGDKSLSGMQQTLDQIKEKFKTTADLFAAVLEVVARR 325
QLKT++QW+AAS A R +++ F +KS+ G+ + ++++ + T +L+ A+ ++RR
Sbjct 478 AQLKTLIQWLAASYAGRSMKFYTFSNKSVDGLGLVISKLRQSYATVGNLYKAIQNALSRR 537
Query 326 GSAFRHSRQWSLWRELL 342
WSLW+ELL
Sbjct 538 SGRL----GWSLWKELL 550
> dre:797792 si:dkey-259k14.2
Length=609
Score = 133 bits (335), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 86/243 (35%), Positives = 122/243 (50%), Gaps = 20/243 (8%)
Query 71 FEVVR-PEIKLENVLQDKLPLLPIQVHLNGDITK-ADGAVQVDFADRFLGGLTMHADHLA 128
FE V P +L +K L + V +G I K G +QVDFA +F+GG + + L
Sbjct 346 FERVSVPASELPKWKSEKKLLKNLHVSADGSIEKEGTGMLQVDFASKFIGGGVLKSG-LV 404
Query 129 QEELIFTLKPELIVVMLFTQALKADEAVVVKGAESFTLYRGYERTFELRAAEVPEGCHWS 188
QEE++F + PELI+ LFT+ L E V + G + ++L GY R+F +
Sbjct 405 QEEILFLMSPELILARLFTEKLDDHECVRITGPQMYSLTSGYSRSFSWTGPYMDR----- 459
Query 189 LHGLCPLDGLGRRAATVAAIDALIVRNPNSQYQENKMKREVLKALLGFQGDPFEALLGEP 248
D RR + AIDAL +NP QY + RE+ KA +GF G+P
Sbjct 460 ----TKRDVWKRRFRQIVAIDALDFKNPLEQYSRENITRELNKAFVGF--------CGQP 507
Query 249 KAPVATGKWGCVIFGGDNQLKTVLQWIAASAAARPLQYMAFGDKSLSGMQQTLDQIKEKF 308
K +ATG WGC F GD +LK +LQ +AA+ R + Y FG+ L+ Q + I +
Sbjct 508 KTAIATGNWGCGAFRGDPKLKALLQLMAAAVVDRDVAYFTFGNTHLANELQKMHDILTQR 567
Query 309 KTT 311
K T
Sbjct 568 KVT 570
> xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase;
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=759
Score = 133 bits (334), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 85/229 (37%), Positives = 120/229 (52%), Gaps = 17/229 (7%)
Query 106 GAVQVDFADRFLGGLTMHADHLAQEELIFTLKPELIVVMLFTQALKADEAVVVKGAESFT 165
G +QVDFA+RF+GG L QEE+ F + PELIV LFT+ L ++E +++ GAE ++
Sbjct 529 GMLQVDFANRFVGGGVTGG-GLVQEEIRFLINPELIVSRLFTEVLDSNECLIITGAEQYS 587
Query 166 LYRGYERTFELRAAEVPEGCHWSLHGLCPLDGLGRRAATVAAIDALIVRNPNSQYQENKM 225
Y GY T++ E P D RR + AIDA R P Q+ K+
Sbjct 588 EYTGYSETYKWACVHEDES---------PRDEWQRRTTEIVAIDAFHFRRPIDQFVPEKI 638
Query 226 KREVLKALLGFQGDPFEALLGEPKAPVATGKWGCVIFGGDNQLKTVLQWIAASAAARPLQ 285
KRE+ KA GF + + + VATG WGC FGGD +LK ++Q +AA+ R L
Sbjct 639 KRELNKAFCGFYR---PEVNPQNLSAVATGNWGCGAFGGDPRLKALIQLLAAAEVGRDLV 695
Query 286 YMAFGDKSL-SGMQQTLDQIKEKFKTTADLFAAVLEV---VARRGSAFR 330
Y FGD+ L + + EK KT D+++ ++E V R S R
Sbjct 696 YFTFGDRELMKDIYLMYSFLTEKNKTVGDIYSMLIEYHNKVCRNCSTPR 744
> ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family
protein
Length=522
Score = 130 bits (326), Expect = 9e-30, Method: Compositional matrix adjust.
Identities = 91/275 (33%), Positives = 134/275 (48%), Gaps = 48/275 (17%)
Query 89 PLLPIQVHLNGDI-TKADGAVQVDFADRFLGGLTMHADHLAQEELIFTLKPELIVVMLFT 147
PL I++H +G I + A++VDFAD + GGLT+ D L QEE+ F + PELI M+F
Sbjct 243 PLCSIEIHTSGAIEDQPCEALEVDFADEYFGGLTLSYDTL-QEEIRFVINPELIAGMIFL 301
Query 148 QALKADEAVVVKGAESFTLYRGYERTFELRAAEVPEGCHWSLHGLCPLDGLGRRAATVAA 207
+ A+EA+ + G E F+ Y GY +F+ LD RR V A
Sbjct 302 PRMDANEAIEIVGVERFSGYTGYGPSFQYAGDYTDNK---------DLDIFRRRKTRVIA 352
Query 208 IDALIVRNPN-SQYQENKMKREVLKALLGF----------QGDPFEALLGEP-------- 248
IDA+ +P QY+ + + REV KA G+ + DP + P
Sbjct 353 IDAM--PDPGMGQYKLDALIREVNKAFSGYMHQCKYNIDVKHDPEASSSHVPLTSDSASQ 410
Query 249 ---------------KAPVATGKWGCVIFGGDNQLKTVLQWIAASAAARP-LQYMAFGDK 292
K VATG WGC +FGGD +LK +LQW+A S + RP + Y FG +
Sbjct 411 VIESSHRWCIDHEEKKIGVATGNWGCGVFGGDPELKIMLQWLAISQSGRPFMSYYTFGLQ 470
Query 293 SLSGMQQTLDQIKEKFKTTADLFAAVLEVVARRGS 327
+L + Q ++ + + T DL+ ++E + R S
Sbjct 471 ALQNLNQVIEMVALQEMTVGDLWKKLVEYSSERLS 505
> dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759
poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=777
Score = 129 bits (325), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 87/231 (37%), Positives = 121/231 (52%), Gaps = 15/231 (6%)
Query 87 KLPLLPIQVHLNGDI-TKADGAVQVDFADRFLGGLTMHADHLAQEELIFTLKPELIVVML 145
K PL + + G I + G +QVDFA+R + G + L QEE+ F + PELIV L
Sbjct 506 KSPLSHLHITCKGTIEDQGYGMLQVDFANR-MVGGGVTGLGLVQEEIRFLINPELIVSRL 564
Query 146 FTQALKADEAVVVKGAESFTLYRGYERTFELRAAEVPEGCHWSLHGLCPLDGLGRRAATV 205
FT+ L +E +++ G E ++ Y GY +F+ E H P DG RR +
Sbjct 565 FTEVLDHNECLIITGTEQYSKYSGYAESFKW------EDNH---KDKIPRDGWQRRCTEI 615
Query 206 AAIDALIVRNPNSQYQENKMKREVLKALLGFQGDPFEALLGEPKAPVATGKWGCVIFGGD 265
A+DAL RN Q+Q KM RE+ KA GF L + VATG WGC FGGD
Sbjct 616 VAMDALHYRNFMDQFQPEKMTRELNKAYCGFMRPGVNPLN---LSAVATGNWGCGAFGGD 672
Query 266 NQLKTVLQWIAASAAARPLQYMAFGDKSLSGMQQTLDQ-IKEKFKTTADLF 315
+LK +LQ +AA+ A R + Y FGD++L Q L + +K+ T L+
Sbjct 673 TRLKALLQLMAAAEAGRDVAYFTFGDEALMRDVQDLHKFLKDNCVTVGSLY 723
> hsa:8505 PARG, FLJ54459, FLJ60257, FLJ60456, PARG99; poly (ADP-ribose)
glycohydrolase (EC:3.2.1.143); K07759 poly(ADP-ribose)
glycohydrolase [EC:3.2.1.143]
Length=976
Score = 117 bits (293), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 81/209 (38%), Positives = 109/209 (52%), Gaps = 18/209 (8%)
Query 89 PLLPIQVHLNGDITK-ADGAVQVDFADRFLGGLTMHADHLAQEELIFTLKPELIVVMLFT 147
PL + V G I + G +QVDFA+RF+GG A L QEE+ F + PELI+ LFT
Sbjct 714 PLTRLHVTYEGTIEENGQGMLQVDFANRFVGGGVTSAG-LVQEEIRFLINPELIISRLFT 772
Query 148 QALKADEAVVVKGAESFTLYRGYERTFELRAAEVPEGCHWSL--HGLCPLDGLGRRAATV 205
+ L +E +++ G E ++ Y GY T+ WS D RR +
Sbjct 773 EVLDHNECLIITGTEQYSEYTGYAETY-----------RWSRSHEDGSERDDWQRRCTEI 821
Query 206 AAIDALIVRNPNSQYQENKMKREVLKALLGFQGDPFEALLGEPKAPVATGKWGCVIFGGD 265
AIDAL R Q+ KM+RE+ KA GF + E + VATG WGC FGGD
Sbjct 822 VAIDALHFRRYLDQFVPEKMRRELNKAYCGFLR---PGVSSENLSAVATGNWGCGAFGGD 878
Query 266 NQLKTVLQWIAASAAARPLQYMAFGDKSL 294
+LK ++Q +AA+AA R + Y FGD L
Sbjct 879 ARLKALIQILAAAAAERDVVYFTFGDSEL 907
> mmu:26430 Parg, AI413217; poly (ADP-ribose) glycohydrolase (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=961
Score = 117 bits (293), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 85/210 (40%), Positives = 113/210 (53%), Gaps = 20/210 (9%)
Query 89 PLLPIQVHLNGDIT-KADGAVQVDFADRFLGGLTMHADHLAQEELIFTLKPELIVVMLFT 147
PL + V G I G +QVDFA+RF+GG A L QEE+ F + PELIV LFT
Sbjct 707 PLTRLHVTYEGTIEGNGRGMLQVDFANRFVGGGVTGAG-LVQEEIRFLINPELIVSRLFT 765
Query 148 QALKADEAVVVKGAESFTLYRGYERTFE-LRAAEVPEGCHWSLHGLCPLDGLGRRAATVA 206
+ L +E +++ G E ++ Y GY T+ R+ E +G D RR +
Sbjct 766 EVLDHNECLIITGTEQYSEYTGYAETYRWARSHE--DGSE--------KDDWQRRCTEIV 815
Query 207 AIDALIVRNPNSQYQENKMKREVLKALLGF--QGDPFEALLGEPKAPVATGKWGCVIFGG 264
AIDAL R Q+ K++RE+ KA GF G P E L + VATG WGC FGG
Sbjct 816 AIDALHFRRYLDQFVPEKVRRELNKAYCGFLRPGVPSENL-----SAVATGNWGCGAFGG 870
Query 265 DNQLKTVLQWIAASAAARPLQYMAFGDKSL 294
D +LK ++Q +AA+AA R + Y FGD L
Sbjct 871 DARLKALIQILAAAAAERDVVYFTFGDSEL 900
> cel:F20C5.1 pme-3; Poly(ADP-ribose) Metabolism Enzyme family
member (pme-3)
Length=764
Score = 106 bits (265), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 82/243 (33%), Positives = 127/243 (52%), Gaps = 23/243 (9%)
Query 87 KLPLLPIQVHLNGDITKADGAV--QVDFADRFLGGLTMHADHLAQEELIFTLKPELIVVM 144
KL LP +V ++ D A+ QVDFA+ LGG ++ + QEE+ F + PE++V M
Sbjct 494 KLRSLP-EVEFFDEMLIEDTALCTQVDFANEHLGGGVLNHGSV-QEEIRFLMCPEMMVGM 551
Query 145 LFTQALKADEAVVVKGAESFTLYRGYERTFELRAAEV-PEGCHWSLHGLCPLDGLGRRAA 203
L + +K EA+ + GA F+ Y GY T L+ AE+ P + + D GR
Sbjct 552 LLCEKMKQLEAISIVGAYVFSSYTGYGHT--LKWAELQPNHSRQNTNEF--RDRFGRLRV 607
Query 204 TVAAIDALIVRNPNSQYQENKMK-----REVLKALLGF--QGDPFEALLGEPKAPVATGK 256
AIDA++ + Q ++ RE+ KA +GF QG F + P+ TG
Sbjct 608 ETIAIDAILFKGSKLDCQTEQLNKANIIREMKKASIGFMSQGPKFTNI------PIVTGW 661
Query 257 WGCVIFGGDNQLKTVLQWIAASAAARPLQYMAFGDKSLSG-MQQTLDQIKEKFKTTADLF 315
WGC F GD LK ++Q IAA A RPL + +FG+ L+ ++ ++++K+K T LF
Sbjct 662 WGCGAFNGDKPLKFIIQVIAAGVADRPLHFCSFGEPELAAKCKKIIERMKQKDVTLGMLF 721
Query 316 AAV 318
+ +
Sbjct 722 SMI 724
> cel:H23L24.5 pme-4; Poly(ADP-ribose) Metabolism Enzyme family
member (pme-4); K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=485
Score = 106 bits (265), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 93/308 (30%), Positives = 141/308 (45%), Gaps = 30/308 (9%)
Query 43 KRHVAAADEVVSFYFQVPEETVFTSPEG---FEVVRPEIK--LENVLQDKLPLLP-IQVH 96
++ A E + F F ++ P G F +R K LEN + LLP +QV
Sbjct 179 QKTTCVAVEKLKFLFTYFDKMSIDPPIGAVSFRKMRITHKQYLENWKLRETNLLPDVQVF 238
Query 97 LNGDITKADGAVQVDFADRFLGGLTMHADHLAQEELIFTLKPELIVVMLFTQALKADEAV 156
I + Q+DFA++ LGG + + QEE+ F + PE++V +L + EA+
Sbjct 239 DKMSIEETALCTQIDFANKRLGGGVLKGGAV-QEEIRFMMCPEMMVAILLNDVTQDLEAI 297
Query 157 VVKGAESFTLYRGYERTFELRAAEVPEGCHWSLHGLCPLDGLGRRAATVAAIDALIVRNP 216
+ GA F+ Y GY T L+ A++ H + + D GR AIDA VRN
Sbjct 298 SIVGAYVFSSYTGYSNT--LKWAKITPK-HSAQNNNSFRDQFGRLQTETVAIDA--VRNA 352
Query 217 NS-------QYQENKMKREVLKALLGF--QGDPFEALLGEPKAPVATGKWGCVIFGGDNQ 267
+ Q K+ REV KA +GF GD F K PV +G WGC F G+
Sbjct 353 GTPLECLLNQLTTEKLTREVRKAAIGFLSAGDGFS------KIPVVSGWWGCGAFRGNKP 406
Query 268 LKTVLQWIAASAAARPLQYMAFGDKSLSGMQQTLDQIKEKFKTTADLFAAVLEVVARRGS 327
LK ++Q IA + RPLQ+ FGD L+ + +++ F+ + ++ G
Sbjct 407 LKFLIQVIACGISDRPLQFCTFGDTELA---KKCEEMMTLFRNNNVRTGQLFLIINSIGP 463
Query 328 AFRHSRQW 335
+S Q+
Sbjct 464 PLNYSEQY 471
> ath:AT2G31870 TEJ; TEJ (Sanskrit for 'bright'); poly(ADP-ribose)
glycohydrolase
Length=547
Score = 77.8 bits (190), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 50/148 (33%), Positives = 77/148 (52%), Gaps = 12/148 (8%)
Query 90 LLPIQVHLNGDI-TKADGAVQVDFADRFLGGLTMHADHLAQEELIFTLKPELIVVMLFTQ 148
L +VH G I + D A++VDFA+++LGG ++ + QEE+ F + PELI MLF
Sbjct 233 LCAFKVHSFGLIEDQPDNALEVDFANKYLGGGSL-SRGCVQEEIRFMINPELIAGMLFLP 291
Query 149 ALKADEAVVVKGAESFTLYRGYERTFELRAAEVPEGCHWSLHGLCPLDGLGRRAATVAAI 208
+ +EA+ + GAE F+ Y GY +F + + +D RR + AI
Sbjct 292 RMDDNEAIEIVGAERFSCYTGYASSFRFAGEYIDKKA---------MDPFKRRRTRIVAI 342
Query 209 DALIVRNPNSQYQENKMKREVLKALLGF 236
DAL +++ + RE+ KAL GF
Sbjct 343 DALCTPK-MRHFKDICLLREINKALCGF 369
Score = 67.0 bits (162), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 33/75 (44%), Positives = 44/75 (58%), Gaps = 1/75 (1%)
Query 252 VATGKWGCVIFGGDNQLKTVLQWIAASAAARP-LQYMAFGDKSLSGMQQTLDQIKEKFKT 310
VATG WGC +FGGD +LK +QW+AAS RP + Y FG ++L + Q I T
Sbjct 446 VATGNWGCGVFGGDPELKATIQWLAASQTRRPFISYYTFGVEALRNLDQVTKWILSHKWT 505
Query 311 TADLFAAVLEVVARR 325
DL+ +LE A+R
Sbjct 506 VGDLWNMMLEYSAQR 520
> tgo:TGME49_062760 poly(ADP-ribose) glycohydrolase, putative
Length=952
Score = 55.5 bits (132), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 31/84 (36%), Positives = 42/84 (50%), Gaps = 3/84 (3%)
Query 213 VRNPNSQYQENKMKREVLKALLGFQGDPFEALLGEPKAPVATGKWGCVIFGGDNQLKTVL 272
V P+ ++N+ + + P + L+ K P ATG WGC +F GD QLK +L
Sbjct 792 VSTPDGALRQNEERDRFSASEATLAPGPHQGLV---KRPFATGNWGCGVFKGDPQLKFLL 848
Query 273 QWIAASAAARPLQYMAFGDKSLSG 296
QW+AAS R L Y A L G
Sbjct 849 QWLAASLVGRRLIYHAHSRPELVG 872
Score = 44.3 bits (103), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 34/103 (33%), Positives = 58/103 (56%), Gaps = 7/103 (6%)
Query 78 IKLENVLQDKLPLLPIQ-VHLNGDITKA-DGAVQV----DFADRFLGGLTMHADHLAQEE 131
+ ++++ + PLLP+Q N + K D AV + DFA++++GG ++ QEE
Sbjct 444 VTMDSLADREDPLLPLQEFRENAEAGKGIDSAVNLEIMADFANQWIGGGALYRG-CVQEE 502
Query 132 LIFTLKPELIVVMLFTQALKADEAVVVKGAESFTLYRGYERTF 174
+ F PEL+++ LF Q L +E+ + GA F+ Y GY +F
Sbjct 503 IFFATHPELLLLRLFQQRLAINESCAMSGAMQFSRYSGYADSF 545
> cpv:cgd8_2160 shares a domain with poly(ADP) ribose glycohydrolases,
some protein kinase A anchoring proteins and baculovirus
HzNV Orf103, possible transmembrane domain within N-terminus
Length=441
Score = 47.8 bits (112), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 57/212 (26%), Positives = 98/212 (46%), Gaps = 24/212 (11%)
Query 107 AVQVDFADRFLGGLTMHADHLAQEELIFTLKPELIVVMLFTQALKADEAVVVKGAESFTL 166
V+ FA++ G T+ + + QEE++ T+ PE ++ F L +++V ++G ++
Sbjct 226 VVEAIFANKVYGSATLQ-NCMNQEEIVMTVVPETLIGRFFLDDLHDEDSVTIRGVMRYSN 284
Query 167 YRGY-ERTFELRAAEVPEGCHWSLHGLCPLDGLGRRAATVAAIDALIVRNPNSQYQENKM 225
Y+G+ F ++ + E ++ P A +DAL + S+++E +
Sbjct 285 YKGFGTDKFTFQSIKESE-----MYMSIP--------RVYAVVDAL---SGGSRFREFTL 328
Query 226 K---REVLKALLGFQGDPFEALLGEPKAPVATGKWGCVIFGGDNQLKTVLQWIAASAAAR 282
RE+ K + D + + TG WG +FGGDNQ K +LQ IAA R
Sbjct 329 DYALREINKLISALCDDFYGESEERDRNQFVTGYWGGGVFGGDNQYKFILQLIAACVCNR 388
Query 283 PLQYMAFGDKSLSGMQQTLDQIKEKFKTTADL 314
L Y + G L Q + QI+ K+ T L
Sbjct 389 QLVY-SDGLNRLDVNQ--IRQIESKYTTCGQL 417
> mmu:11642 Akap3, Prka3; A kinase (PRKA) anchor protein 3
Length=864
Score = 36.2 bits (82), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 45/97 (46%), Gaps = 10/97 (10%)
Query 233 LLGFQGDPFEALLGEPKAPVATGKWGCVIFGGDNQLKTVLQWIAASAAARPLQYMAFGDK 292
L G G P E PK V+ + + QL+ VLQW+AAS P+ Y A D+
Sbjct 752 LRGLSGQPPEGC-EIPKVIVSNHNLADTV--QNKQLQAVLQWVAASELNVPILYFAGDDE 808
Query 293 SLSGMQQTLDQIK----EKFKTTADLFAAVLEVVARR 325
G+Q+ L Q+ EK ++ ++ +VL R
Sbjct 809 ---GIQEKLLQLSATAVEKGRSVGEVLQSVLRYEKER 842
> mmu:11643 Akap4, Fsc1, p82; A kinase (PRKA) anchor protein 4
Length=840
Score = 33.9 bits (76), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 30/68 (44%), Gaps = 1/68 (1%)
Query 259 CVIFGGDNQLKTVLQWIAASAAARPLQY-MAFGDKSLSGMQQTLDQIKEKFKTTADLFAA 317
C QL+ VLQWIAAS P+ Y M D L + + + EK + DL
Sbjct 751 CSTTNLQKQLQAVLQWIAASQFNVPMLYFMGDDDGQLEKLPEVSAKAAEKGYSVGDLLQE 810
Query 318 VLEVVARR 325
V++ R
Sbjct 811 VMKFAKER 818
> mmu:100503160 a-kinase anchor protein 4-like
Length=712
Score = 32.3 bits (72), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 29/63 (46%), Gaps = 1/63 (1%)
Query 259 CVIFGGDNQLKTVLQWIAASAAARPLQY-MAFGDKSLSGMQQTLDQIKEKFKTTADLFAA 317
C QL+ VLQWIAAS P+ Y M D L + + + EK + DL
Sbjct 623 CSTSNLQKQLQAVLQWIAASQFNVPMLYFMGDDDGQLEKLPEVSAKAAEKGYSVGDLLQE 682
Query 318 VLE 320
V++
Sbjct 683 VMK 685
> hsa:10566 AKAP3, AKAP110, CT82, FSP95, PRKA3, SOB1; A kinase
(PRKA) anchor protein 3
Length=853
Score = 32.3 bits (72), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 25/40 (62%), Gaps = 3/40 (7%)
Query 265 DNQLKTVLQWIAASAAARPLQYMAFGDKSLSGMQQTLDQI 304
+ QL+ VLQW+AAS P+ Y A D+ G+Q+ L Q+
Sbjct 770 NKQLQAVLQWVAASELNVPILYFAGDDE---GIQEKLLQL 806
> mmu:77629 Sphkap, 4930544G21Rik, A930009L15Rik, AI852220, mKIAA1678;
SPHK1 interactor, AKAP domain containing
Length=1658
Score = 31.6 bits (70), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 41/83 (49%), Gaps = 7/83 (8%)
Query 265 DNQLKTVLQWIAASAAARPLQYMAFGDKSLSGMQQTLDQIK---EKFKTTADLFAAVLEV 321
D +L+ LQWIAAS P Y F S +++ LD +K +K D+F AV++
Sbjct 1579 DAELRATLQWIAASELGIPTIY--FKKSQESRIEKFLDVVKLVQQKSWKVGDIFHAVVQY 1636
Query 322 VARRGSAFRHSRQWSLWRELLSL 344
+ A + R SL+ LL L
Sbjct 1637 C--KLHAEQKERTPSLFDWLLEL 1657
> hsa:8852 AKAP4, AKAP_82, AKAP-4, AKAP82, CT99, FSC1, HI, PRKA4,
hAKAP82, p82; A kinase (PRKA) anchor protein 4
Length=854
Score = 31.2 bits (69), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 29/68 (42%), Gaps = 1/68 (1%)
Query 259 CVIFGGDNQLKTVLQWIAASAAARPLQY-MAFGDKSLSGMQQTLDQIKEKFKTTADLFAA 317
C QL+ VLQWIAAS P+ Y M D L + Q + EK + L
Sbjct 765 CSTNSLQKQLQAVLQWIAASQFNVPMLYFMGDKDGQLEKLPQVSAKAAEKGYSVGGLLQE 824
Query 318 VLEVVARR 325
V++ R
Sbjct 825 VMKFAKER 832
> mmu:219103 Cenpj, 4932437H03Rik, CPAP, Gm81, LAP, LIP1, MGC102215;
centromere protein J; K11502 centromere protein J
Length=1344
Score = 30.4 bits (67), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 34/124 (27%), Positives = 48/124 (38%), Gaps = 28/124 (22%)
Query 14 GWRAVNSSSSSSSSSSSGCRCTEVTFLGTKRHVAAADEVVSFYFQVPEETVFTSPEGFEV 73
G R V + + S+ + VTF DE V +Y+ + T T PEG EV
Sbjct 1173 GRRVVLFPNGTRKEVSADGKSVTVTFFNGDVKQVMPDERVVYYYAAAQTTHTTYPEGLEV 1232
Query 74 VRPEIKLENVLQDKLPLLPIQVHL-NGDITK--ADGAVQVDFADRFLGGLTMHADHLAQE 130
+H +G I K DG ++ F D+ + T+ AD QE
Sbjct 1233 ---------------------LHFSSGQIEKHFPDGRKEITFPDQTIK--TLFAD--GQE 1267
Query 131 ELIF 134
E IF
Sbjct 1268 ESIF 1271
> dre:100329772 GTPase, IMAP family member 4-like
Length=673
Score = 30.4 bits (67), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 292 KSLSGMQQTLDQIKEKFKTTADLFAAVLEVVARRGSA 328
K + +Q LD+ K+K TAD++AA ++ + R +A
Sbjct 621 KEMEDCRQILDEAMRKYKETADMYAAEVKRIEARNAA 657
Lambda K H
0.319 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 14664651540
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40