bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2249_orf1
Length=223
Score E
Sequences producing significant alignments: (Bits) Value
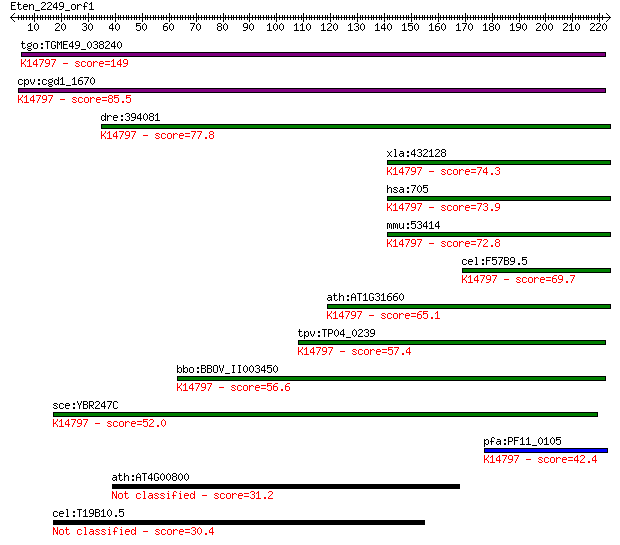
tgo:TGME49_038240 bystin, putative (EC:3.4.24.61); K14797 esse... 149 9e-36
cpv:cgd1_1670 bystin/ S.cerevisiae En1p like adaptor domain ; ... 85.5 1e-16
dre:394081 bysl, MGC63889, zgc:63889; bystin-like; K14797 esse... 77.8 3e-14
xla:432128 bysl, MGC81422; bystin-like; K14797 essential nucle... 74.3 3e-13
hsa:705 BYSL, BYSTIN; bystin-like; K14797 essential nuclear pr... 73.9 4e-13
mmu:53414 Bysl, Bys, Enp1; bystin-like; K14797 essential nucle... 72.8 8e-13
cel:F57B9.5 byn-1; mammalian BYstiN (adhesion protein) related... 69.7 8e-12
ath:AT1G31660 hypothetical protein; K14797 essential nuclear p... 65.1 2e-10
tpv:TP04_0239 hypothetical protein; K14797 essential nuclear p... 57.4 4e-08
bbo:BBOV_II003450 18.m06289; bystin family protein; K14797 ess... 56.6 6e-08
sce:YBR247C ENP1, MEG1; Enp1p; K14797 essential nuclear protein 1 52.0
pfa:PF11_0105 rRNA processing protein, putative; K14797 essent... 42.4 0.001
ath:AT4G00800 binding / protein binding / zinc ion binding 31.2 3.0
cel:T19B10.5 hypothetical protein 30.4 5.5
> tgo:TGME49_038240 bystin, putative (EC:3.4.24.61); K14797 essential
nuclear protein 1
Length=558
Score = 149 bits (375), Expect = 9e-36, Method: Compositional matrix adjust.
Identities = 98/236 (41%), Positives = 139/236 (58%), Gaps = 26/236 (11%)
Query 5 PKPERRRRKGAPRHNPLYEDVLDARGIPLPGGAIDAPRNVKIKQAVEDAEAVENCTRDDQ 64
PK + +RK A RHNPL+ D+L G+ P + + K K A + + + +
Sbjct 86 PKVQSEKRKVANRHNPLHVDILGQAGVHTPETGLLFQK--KGKAAKKSRLSRSADDDEGE 143
Query 65 EQEGLLPPKLGKKLLKLVEEQQ--RAEGIADAS----------DAEGGGPDEETADKDGF 112
+ +G++P +L KK+L+LVEEQQ R+E DA D + GG D D +GF
Sbjct 144 DADGVVPRRLSKKILRLVEEQQHHRSENGPDADSGSELDSEDVDRDSGGED---VDSEGF 200
Query 113 VVLDCEEDEEDLVFEK---QRTGSSATRATVNLADLVMEQLKKQTENKDKSQEKRVE--- 166
VV+D E DEED ++ + QR G++A T LAD ++E+L+++ E +
Sbjct 201 VVIDGEADEEDELYVQRVQQRQGTAAAAPT--LADFILEKLRQKEERESSGAAAEAPEED 258
Query 167 -SSLSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTMWSKQA 221
S+L PKVVEVYTAM FL +YRSGKMPKAFK++PRL WEE+L+LT P WS+QA
Sbjct 259 CSALPPKVVEVYTAMGSFLQKYRSGKMPKAFKVLPRLQRWEEVLLLTQPESWSRQA 314
> cpv:cgd1_1670 bystin/ S.cerevisiae En1p like adaptor domain
; K14797 essential nuclear protein 1
Length=461
Score = 85.5 bits (210), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 66/241 (27%), Positives = 111/241 (46%), Gaps = 31/241 (12%)
Query 4 MPKPERRRRKGAPRHNPLYEDVLDARGIPLPGGAIDAPRNVKIKQAVEDAEAVENCTRDD 63
MPK ++ + K + NPL+ D+ A PR +K + D+ +N + ++
Sbjct 1 MPKFKKSKNKPSRNRNPLHVDIESA------SCPYSKPRKIKPIDSSHDS-LYDNLSNEE 53
Query 64 QEQEGLLPPKLGKKLLKLVEEQ-------QRAEGIADASDAEGGGPDEETADKDGFVVLD 116
Q + P +GKK+L Q + E I E D GF+ +D
Sbjct 54 QFSSEV-PKNIGKKILNEANRQLNDFYDDEIQEHIIYEETNESEEKSNLPIDSAGFIYID 112
Query 117 -----CEEDE-------EDLV--FEKQRTGSSATRATVNLADLVMEQLKKQTENKD--KS 160
+D DL+ +E + S + R D ++ +L++++ ++ +
Sbjct 113 ESGIGTADDMGLDNSYWNDLLKNYENENHSSQSIRNQTGFIDTIVSKLREKSSDRKPLEI 172
Query 161 QEKRVESSLSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTMWSKQ 220
E S++ KV +VYT + +L +Y+SGK+PKAF IIP+L WEE++ LTNP WS
Sbjct 173 SESCQNSNIPEKVTQVYTLIGEWLTKYKSGKLPKAFSIIPKLENWEEVVFLTNPHNWSPN 232
Query 221 A 221
A
Sbjct 233 A 233
> dre:394081 bysl, MGC63889, zgc:63889; bystin-like; K14797 essential
nuclear protein 1
Length=422
Score = 77.8 bits (190), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 69/215 (32%), Positives = 105/215 (48%), Gaps = 32/215 (14%)
Query 35 GGAIDAPRNVKIKQAVEDAEAVENCTRDDQEQEGLLPPKLGKKLLKLVEEQQRAEGIADA 94
GGA+ + + +V + + R +EQ+ + KL +++L+ QQ E + D
Sbjct 10 GGAVPLAEQILQQDSVRCSGRQKQRGRGQEEQQEFVDEKLSRRILEQARRQQ--EELQDT 67
Query 95 SD------AEGGGPDEETADKD------GFVVLDCE-------EDEEDLVFEKQRTGSSA 135
++ A GP + D D G + + E EDE+ + Q S
Sbjct 68 AEEERRTPATRLGPVSQDGDSDEEWPALGEAIDEVEPEVEVDPEDEQAI----QMFMSKN 123
Query 136 TRATVNLADLVMEQL-KKQTENKDKSQEKRVES--SLSPKVVEVYTAMAPFLARYRSGKM 192
LAD++ME++ +KQTE E + + P+VVEVY ++ L++YRSGK+
Sbjct 124 PPMRRTLADIIMEKITEKQTEVGTVLSEVSGHAMPQMDPRVVEVYRGVSKVLSKYRSGKL 183
Query 193 PKAFKIIPRLHAWEEILVLTNPTMWSK----QATR 223
PKAFKIIP L WE++L LT P WS QATR
Sbjct 184 PKAFKIIPALSNWEQVLYLTEPETWSAAAMYQATR 218
> xla:432128 bysl, MGC81422; bystin-like; K14797 essential nuclear
protein 1
Length=431
Score = 74.3 bits (181), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 41/90 (45%), Positives = 57/90 (63%), Gaps = 7/90 (7%)
Query 141 NLADLVMEQL-KKQTENKDKSQE--KRVESSLSPKVVEVYTAMAPFLARYRSGKMPKAFK 197
LAD++ME++ +KQTE + E R L P+++EVY + L+ YRSGK+PKAFK
Sbjct 143 TLADIIMEKITEKQTEVETMMSEVSGRPMPQLDPRILEVYKGVKEVLSSYRSGKLPKAFK 202
Query 198 IIPRLHAWEEILVLTNPTMWS----KQATR 223
I+P L WE+IL +T P W+ QATR
Sbjct 203 IVPALSNWEQILYITEPEAWTAAAVYQATR 232
> hsa:705 BYSL, BYSTIN; bystin-like; K14797 essential nuclear
protein 1
Length=437
Score = 73.9 bits (180), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 43/90 (47%), Positives = 57/90 (63%), Gaps = 7/90 (7%)
Query 141 NLADLVMEQL-KKQTENKDKSQEKR--VESSLSPKVVEVYTAMAPFLARYRSGKMPKAFK 197
LAD++ME+L +KQTE + E L P+V+EVY + L++YRSGK+PKAFK
Sbjct 146 TLADIIMEKLTEKQTEVETVMSEVSGFPMPQLDPRVLEVYRGVREVLSKYRSGKLPKAFK 205
Query 198 IIPRLHAWEEILVLTNPTMWSK----QATR 223
IIP L WE+IL +T P W+ QATR
Sbjct 206 IIPALSNWEQILYVTEPEAWTAAAMYQATR 235
> mmu:53414 Bysl, Bys, Enp1; bystin-like; K14797 essential nuclear
protein 1
Length=436
Score = 72.8 bits (177), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 42/90 (46%), Positives = 56/90 (62%), Gaps = 7/90 (7%)
Query 141 NLADLVMEQL-KKQTENKDKSQEKR--VESSLSPKVVEVYTAMAPFLARYRSGKMPKAFK 197
LAD++ME+L +KQTE + E L P+V+EVY + L +YRSGK+PKAFK
Sbjct 145 TLADIIMEKLTEKQTEVETVMSEVSGFPMPQLDPRVLEVYRGVREVLCKYRSGKLPKAFK 204
Query 198 IIPRLHAWEEILVLTNPTMWSK----QATR 223
+IP L WE+IL +T P W+ QATR
Sbjct 205 VIPALSNWEQILYVTEPEAWTAAAMYQATR 234
> cel:F57B9.5 byn-1; mammalian BYstiN (adhesion protein) related
family member (byn-1); K14797 essential nuclear protein 1
Length=449
Score = 69.7 bits (169), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 30/59 (50%), Positives = 43/59 (72%), Gaps = 4/59 (6%)
Query 169 LSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTMWS----KQATR 223
+ P+VVE+Y + ++++YRSGK+PKAFKIIP++ WE+IL LT P W+ QATR
Sbjct 179 MDPEVVEMYEQIGQYMSKYRSGKVPKAFKIIPKMINWEQILFLTKPETWTAAAMYQATR 237
> ath:AT1G31660 hypothetical protein; K14797 essential nuclear
protein 1
Length=444
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 65/109 (59%), Gaps = 9/109 (8%)
Query 119 EDEEDLVFEKQRTGSSATRATVNLADLVMEQLKKQTENKDKSQEKRVESSLSPKVVEVYT 178
ED+E L FE ++ + T L D+++++LK + + D ++E+R + + P + ++Y
Sbjct 118 EDDEKL-FESFLNKNAPPQRT--LTDIIIKKLKDK--DADLAEEERPDPKMDPAITKLYK 172
Query 179 AMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTMWS----KQATR 223
+ F++ Y GK+PKAFK++ + WE++L LT P WS QATR
Sbjct 173 GVGKFMSEYTVGKLPKAFKLVTSMEHWEDVLYLTEPEKWSPNALYQATR 221
> tpv:TP04_0239 hypothetical protein; K14797 essential nuclear
protein 1
Length=384
Score = 57.4 bits (137), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 55/114 (48%), Gaps = 17/114 (14%)
Query 108 DKDGFVVLDCEEDEEDLVFEKQRTGSSATRATVNLADLVMEQLKKQTENKDKSQEKRVES 167
+ D V D ED + E Q T +++ E K + K +E V
Sbjct 86 ENDPLVSFDFSEDSAGFLPESQTTSNTS------------EIWSKLNDLSTKPKES-VMD 132
Query 168 SLSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTMWSKQA 221
L P VY + +L+RY+SG +PKAFK++P++ W EI+ LTNP W+ A
Sbjct 133 RLRP----VYNEIGLYLSRYKSGGLPKAFKVLPKMTNWAEIIQLTNPQNWTPNA 182
> bbo:BBOV_II003450 18.m06289; bystin family protein; K14797 essential
nuclear protein 1
Length=383
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 49/166 (29%), Positives = 77/166 (46%), Gaps = 34/166 (20%)
Query 63 DQEQEGL--LPPKLGKKLLKLVEEQ-QRAEGIADASDAEGGGPDEETADK----DGFVVL 115
D E E L +P + KK+ K+V E Q + IA +D++ D+ +D DGF+
Sbjct 44 DIEAEYLDDIPEFIAKKVHKMVNENDQESIKIAIVNDSDFLSQDKLVSDTLNEIDGFM-- 101
Query 116 DCEEDEEDLVFEKQRTGSSATRATVNLADLVMEQLKKQTENKDKSQEKRVESSLSPKVVE 175
SS +T+ + L +E K E ES K+
Sbjct 102 ---------------PESSIPSSTIEIGQL--------SERIFKIPEAETESI--RKMRA 136
Query 176 VYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTMWSKQA 221
V++ + ++++Y SG +PKAFK +PR+ WEE L LT+P W+ A
Sbjct 137 VFSEIGVYMSKYTSGGLPKAFKFLPRMSNWEEFLELTHPENWTPNA 182
> sce:YBR247C ENP1, MEG1; Enp1p; K14797 essential nuclear protein
1
Length=483
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 59/258 (22%), Positives = 103/258 (39%), Gaps = 71/258 (27%)
Query 17 RHNPLYEDVLDARGIPLPGGAIDAPRNVKIKQAVEDAEAVENCTRDDQEQEGLLPPKLGK 76
RH+PL +D+ A+G + + K+ ++ A + E++G + K +
Sbjct 13 RHDPLLKDLDAAQG---------TLKKINKKKLAQNDAANHDAA---NEEDGYIDSKASR 60
Query 77 KLLKLVEEQQ---RAEGIA---------------------------------DASDAEGG 100
K+L+L +EQQ E +A D SD E
Sbjct 61 KILQLAKEQQDEIEGEELAESERNKQFEARFTTMSYDDEDEDEDEDEEAFGEDISDFEPE 120
Query 101 GPDEETADKDGFVVLDCEEDEEDLVFEKQRTGSSATRATVNLADLVMEQLKK---QTE-- 155
G +E + D E+ + K+ ++ + NLAD +M +++ Q E
Sbjct 121 GDYKEEEEIVEIDEEDAAMFEQ---YFKKSDDFNSLSGSYNLADKIMASIREKESQVEDM 177
Query 156 -------NKDKSQEKRVESSLSP--------KVVEVYTAMAPFLARYRSGKMPKAFKIIP 200
N+ + + S L KV++ YT + L + GK+PK FK+IP
Sbjct 178 QDDEPLANEQNTSRGNISSGLKSGEGVALPEKVIKAYTTVGSILKTWTHGKLPKLFKVIP 237
Query 201 RLHAWEEILVLTNPTMWS 218
L W++++ +TNP WS
Sbjct 238 SLRNWQDVIYVTNPEEWS 255
> pfa:PF11_0105 rRNA processing protein, putative; K14797 essential
nuclear protein 1
Length=436
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 177 YTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTMWSKQAT 222
Y + LA Y+SGK+ +A I+ + W E L+LT P W+ AT
Sbjct 179 YRTIGEDLAHYKSGKLHRALTILTKSSKWYEYLLLTKPKKWTAHAT 224
> ath:AT4G00800 binding / protein binding / zinc ion binding
Length=1918
Score = 31.2 bits (69), Expect = 3.0, Method: Composition-based stats.
Identities = 31/135 (22%), Positives = 61/135 (45%), Gaps = 18/135 (13%)
Query 39 DAPRNVKIKQAVEDAEAVENCTRDDQEQEGLLPPKLGKKLLKLVEEQQRAEGIADASDAE 98
D+ NV ++ E N DD E +L + LVE + +D+E
Sbjct 235 DSEGNVSTEKEAETTMEAGNAAIDDDTDETML-------VASLVESSESQH----LTDSE 283
Query 99 GGGPDEETADKDGFVVLDCEEDEEDLVF-EKQRTGSSA-----TRATVNLADLVMEQLKK 152
G D + ++ + V D + D+ D++ E ++ G A + +++LV E++
Sbjct 284 GKCDDAKVSNDEESSVGDVKSDKSDIIIPESKKEGGDAFIPDDGSSMSGISELVEERI-A 342
Query 153 QTENKDKSQEKRVES 167
+ EN+ S+ +R++S
Sbjct 343 ELENERMSKRERLKS 357
> cel:T19B10.5 hypothetical protein
Length=705
Score = 30.4 bits (67), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 32/142 (22%), Positives = 59/142 (41%), Gaps = 27/142 (19%)
Query 17 RHNPLYEDVLDARGIPLPGGAIDAPRNVKIKQAVEDAEAVENCTRDDQEQEGLLPPKLGK 76
R++PL+ P G I P ++ A + CT D +Q+ LPP+ +
Sbjct 334 RYHPLFRSNTLTHVHFNPLGTISTPTSI--------ATTPQRCTSTDSKQKHALPPQ--R 383
Query 77 KLLKLVEEQQRAEGIADASDAEGGGPDEETADKDGFVVLDCEEDEEDLVFEKQRTGSSAT 136
++++ + A+ + A DA+ D DG + DC E T S T
Sbjct 384 EIIE-PDSTTCADDNSQARDADNSRMDSSDYGSDG--ISDCRES----------TASDGT 430
Query 137 RAT----VNLADLVMEQLKKQT 154
+ T + + D++ +++K T
Sbjct 431 QKTLSDRIEVTDIIFDKVKSPT 452
Lambda K H
0.311 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7395169300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40