bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2364_orf1
Length=150
Score E
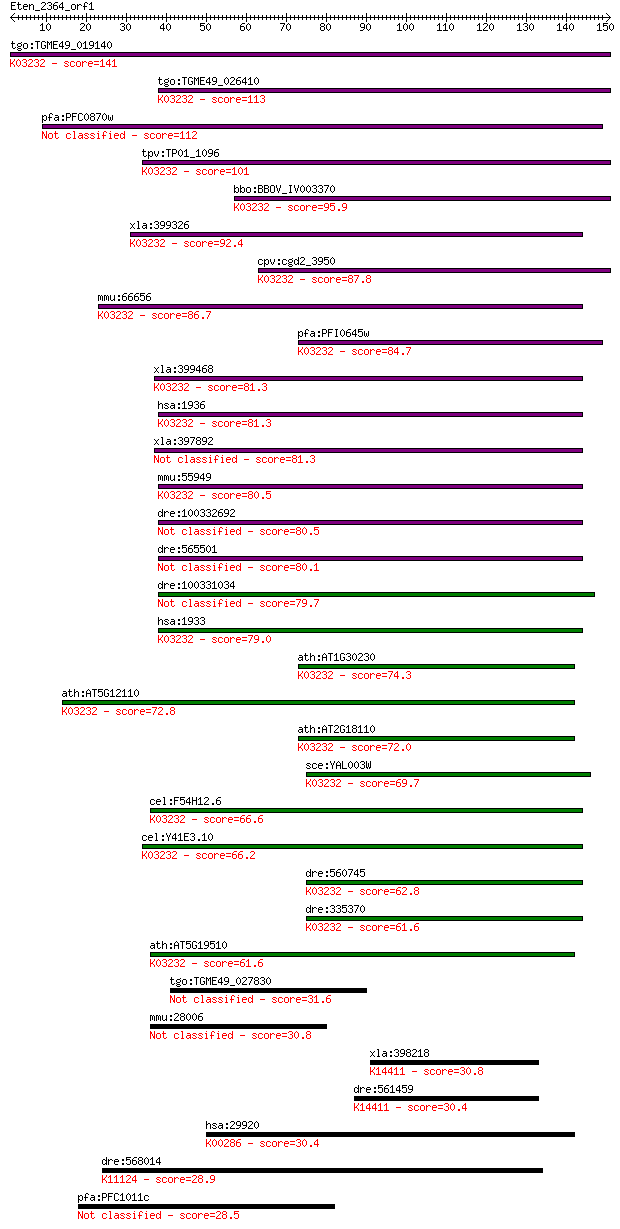
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_019140 elongation factor 1, putative ; K03232 elong... 141 7e-34
tgo:TGME49_026410 elongation factor 1-beta, putative ; K03232 ... 113 2e-25
pfa:PFC0870w elongation factor 1 (EF-1), putative 112 4e-25
tpv:TP01_1096 elongation factor 1 beta; K03232 elongation fact... 101 1e-21
bbo:BBOV_IV003370 21.m02924; translation elongation factor-1 b... 95.9 4e-20
xla:399326 eef1b2, MGC130878, eef1b, ef1b, wu:fj06d02; eukaryo... 92.4 5e-19
cpv:cgd2_3950 translation elongation factor 1 beta 1 ; K03232 ... 87.8 1e-17
mmu:66656 Eef1d, 1700026P12Rik, 5730529A16Rik, AL023999; eukar... 86.7 2e-17
pfa:PFI0645w PfEF-1beta; elongation factor 1-beta; K03232 elon... 84.7 1e-16
xla:399468 eef1d; elongation factor-1 delta; K03232 elongation... 81.3 1e-15
hsa:1936 EEF1D, EF-1D, EF1D, FLJ20897, FP1047; eukaryotic tran... 81.3 1e-15
xla:397892 eef1d; eukaryotic translation elongation factor 1 d... 81.3 1e-15
mmu:55949 Eef1b2, 2810017J07Rik, Eef1b; eukaryotic translation... 80.5 2e-15
dre:100332692 elongation factor-1, delta-like 80.5 2e-15
dre:565501 eef1da, MGC111981, MGC55300, eef1d, zgc:66406; elon... 80.1 2e-15
dre:100331034 eef1d; elongation factor-1, delta-like 79.7 3e-15
hsa:1933 EEF1B2, EEF1B, EEF1B1, EF1B; eukaryotic translation e... 79.0 6e-15
ath:AT1G30230 elongation factor 1-beta / EF-1-beta; K03232 elo... 74.3 1e-13
ath:AT5G12110 elongation factor 1B alpha-subunit 1 (eEF1Balpha... 72.8 4e-13
ath:AT2G18110 elongation factor 1-beta, putative / EF-1-beta, ... 72.0 6e-13
sce:YAL003W EFB1, TEF5; Efb1p; K03232 elongation factor 1-beta 69.7 3e-12
cel:F54H12.6 hypothetical protein; K03232 elongation factor 1-... 66.6 3e-11
cel:Y41E3.10 hypothetical protein; K03232 elongation factor 1-... 66.2 4e-11
dre:560745 eef1db, MGC114052, eef1d, im:7233939, si:dkey-235d1... 62.8 4e-10
dre:335370 eef1b2, MGC86802, fj06d02, wu:fj06d02, zgc:56277, z... 61.6 9e-10
ath:AT5G19510 elongation factor 1B alpha-subunit 2 (eEF1Balpha... 61.6 1e-09
tgo:TGME49_027830 mitochondrial import inner membrane transloc... 31.6 0.92
mmu:28006 D6Wsu116e, A130095H06, C530005J20Rik, Fam21; DNA seg... 30.8 1.6
xla:398218 dazap1; DAZ associated protein 1; K14411 RNA-bindin... 30.8 1.8
dre:561459 dazap1; DAZ associated protein 1; K14411 RNA-bindin... 30.4 2.0
hsa:29920 PYCR2, FLJ54750, P5CR2; pyrroline-5-carboxylate redu... 30.4 2.0
dre:568014 mKIAA0732 protein-like; K11124 protein SMG6 [EC:3.1... 28.9 6.9
pfa:PFC1011c conserved Plasmodium membrane protein, unknown fu... 28.5 7.9
> tgo:TGME49_019140 elongation factor 1, putative ; K03232 elongation
factor 1-beta
Length=187
Score = 141 bits (356), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 70/153 (45%), Positives = 111/153 (72%), Gaps = 6/153 (3%)
Query 1 LTMPLSAENELYVPCYYYIHSQKESGKHATD---DSDDIDLFGDDAEDSAAALKQLSAAK 57
+ + +EL PC YY + ++ D DD DLFG+++ + A+K+L+ +K
Sbjct 1 MVTAFATSDELATPCTYYTLTWGDAAAAPKKAAVDDDDFDLFGEESAEDKEAVKKLAESK 60
Query 58 QQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGNEIKQIQIEGLTWGENQKKVPLA 117
+++ ++++ + V+NKSMLVIEVKP DA+TDL+++ ++K IQ+EG+TWGE KKVP+A
Sbjct 61 KKEAEKKK---KVVINKSMLVIEVKPADADTDLDDVCKKVKSIQMEGVTWGEGMKKVPVA 117
Query 118 FGLYKLQVQCVIVDDLVNTDTVIERIEEIGLSE 150
FGL+KLQVQCVI+DD+VNT+ +++ IEEIG++E
Sbjct 118 FGLFKLQVQCVILDDVVNTNALVDEIEEIGMTE 150
> tgo:TGME49_026410 elongation factor 1-beta, putative ; K03232
elongation factor 1-beta
Length=328
Score = 113 bits (283), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 60/113 (53%), Positives = 88/113 (77%), Gaps = 3/113 (2%)
Query 38 LFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGNEI 97
LFG+D D A +K+L A+ ++++ K+K+ V+NKS LVIEVKP DAET L+EI
Sbjct 181 LFGEDDADKEA-VKKL--AESKKKEAAGKKKKEVINKSSLVIEVKPADAETSLDEISKLC 237
Query 98 KQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIERIEEIGLSE 150
K+I+IEG+TWGE KKVP+AFGLYKLQ+ C I+DD+VNT+ ++++IE +G+++
Sbjct 238 KEIKIEGVTWGEAVKKVPVAFGLYKLQLCCTILDDIVNTNEIVDQIEALGMTQ 290
> pfa:PFC0870w elongation factor 1 (EF-1), putative
Length=156
Score = 112 bits (281), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 62/140 (44%), Positives = 81/140 (57%), Gaps = 28/140 (20%)
Query 9 NELYVPCYYYIHSQKESGKHATDDSDDIDLFGDDAEDSAAALKQLSAAKQQQQQQQRKQK 68
+ELYVP YYI Q E G + +K Q ++ +K
Sbjct 6 DELYVPLSYYIL-QNEGG---------------------------NTSKIDQANTKKPKK 37
Query 69 QPVVNKSMLVIEVKPNDAETDLNEIGNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCV 128
+ V+NKS L+I++KP TDL+E+ +K I +EGLTWG+ KK P AFGL+KLQV CV
Sbjct 38 KEVINKSSLIIDIKPYGENTDLDEVLKLVKNITMEGLTWGKAHKKTPFAFGLFKLQVSCV 97
Query 129 IVDDLVNTDTVIERIEEIGL 148
IVDDLVNTD +IE IE +GL
Sbjct 98 IVDDLVNTDELIETIENLGL 117
> tpv:TP01_1096 elongation factor 1 beta; K03232 elongation factor
1-beta
Length=238
Score = 101 bits (251), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 57/117 (48%), Positives = 78/117 (66%), Gaps = 5/117 (4%)
Query 34 DDIDLFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEI 93
DD+DLFG+ E+ +LK K+ + + K K+ V KS LVI V+P + DL+E+
Sbjct 92 DDLDLFGEADEEEDDSLK-----KKMEAMKAAKTKKKEVAKSSLVIHVEPASVDVDLDEV 146
Query 94 GNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIERIEEIGLSE 150
++ ++IEGLTWGE ++PLAFG+ KLQV C IVDDLVNT+ V E IE +GLSE
Sbjct 147 LKLVRSLKIEGLTWGEASTRIPLAFGIEKLQVMCTIVDDLVNTNEVTEMIENLGLSE 203
> bbo:BBOV_IV003370 21.m02924; translation elongation factor-1
beta subunit; K03232 elongation factor 1-beta
Length=240
Score = 95.9 bits (237), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 44/94 (46%), Positives = 66/94 (70%), Gaps = 0/94 (0%)
Query 57 KQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGNEIKQIQIEGLTWGENQKKVPL 116
K+ + + K K+ KS LVI ++P +TDL+E+ +K+I++EG+TWG K+PL
Sbjct 108 KKMEAMKASKGKKREAAKSSLVIHIEPASVDTDLDEVLRLVKEIKLEGVTWGAASAKIPL 167
Query 117 AFGLYKLQVQCVIVDDLVNTDTVIERIEEIGLSE 150
A+G+ KLQV C I+DDLVNT+ + E IEE+GL+E
Sbjct 168 AYGIQKLQVSCTILDDLVNTNEITELIEELGLTE 201
> xla:399326 eef1b2, MGC130878, eef1b, ef1b, wu:fj06d02; eukaryotic
translation elongation factor 1 beta 2; K03232 elongation
factor 1-beta
Length=227
Score = 92.4 bits (228), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 45/113 (39%), Positives = 76/113 (67%), Gaps = 1/113 (0%)
Query 31 DDSDDIDLFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDL 90
+D DDIDLFG D E+ + K++ + Q + ++ +K ++ KS ++++VKP D ETD+
Sbjct 98 EDDDDIDLFGSDDEEESEDAKRVRDERLAQYEAKKSKKPTLIAKSSILLDVKPWDDETDM 157
Query 91 NEIGNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIERI 143
++ ++ IQ++GL WG + K VP+ +G+ KLQ+QCV+ DD V TD + E+I
Sbjct 158 GKLEECLRSIQMDGLLWG-SSKLVPVGYGIKKLQIQCVVEDDKVGTDVLEEKI 209
> cpv:cgd2_3950 translation elongation factor 1 beta 1 ; K03232
elongation factor 1-beta
Length=247
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 37/88 (42%), Positives = 64/88 (72%), Gaps = 0/88 (0%)
Query 63 QQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGNEIKQIQIEGLTWGENQKKVPLAFGLYK 122
Q +K K+ +KS LV+++KP+ + DL+ + I+ ++IEG+ + E +KKVP+AFGL+K
Sbjct 122 QDKKAKEKPASKSSLVLDIKPSSLDVDLDAVAKMIRALKIEGVEFSEGEKKVPVAFGLFK 181
Query 123 LQVQCVIVDDLVNTDTVIERIEEIGLSE 150
LQ+ I+DDLVNT +++ IE +G+++
Sbjct 182 LQMGATIIDDLVNTQDIVDSIETLGMTD 209
> mmu:66656 Eef1d, 1700026P12Rik, 5730529A16Rik, AL023999; eukaryotic
translation elongation factor 1 delta (guanine nucleotide
exchange protein); K03232 elongation factor 1-beta
Length=281
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 49/122 (40%), Positives = 77/122 (63%), Gaps = 2/122 (1%)
Query 23 KESGKHATDDSD-DIDLFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEV 81
K+ A DD D DIDLFG D E+ +L + +Q +++ +K +V KS ++++V
Sbjct 143 KKGATPAEDDEDKDIDLFGSDEEEEDKEAARLREERLRQYAEKKAKKPTLVAKSSILLDV 202
Query 82 KPNDAETDLNEIGNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIE 141
KP D ETD+ ++ ++ IQ++GL WG + K VP+ +G+ KLQ+QCV+ DD V TD + E
Sbjct 203 KPWDDETDMAQLETCVRSIQLDGLVWGAS-KLVPVGYGIRKLQIQCVVEDDKVGTDLLEE 261
Query 142 RI 143
I
Sbjct 262 EI 263
> pfa:PFI0645w PfEF-1beta; elongation factor 1-beta; K03232 elongation
factor 1-beta
Length=276
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 40/77 (51%), Positives = 56/77 (72%), Gaps = 1/77 (1%)
Query 73 NKSMLVIEVKPNDAETDLNEIGNEIKQ-IQIEGLTWGENQKKVPLAFGLYKLQVQCVIVD 131
N+S+L+IE+KP +TD+ +I +KQ I E + WGE KK+P+AFGLYKL + C+I D
Sbjct 152 NRSILIIEIKPKSIDTDIAKIPKLVKQKIVDENIKWGEEVKKLPVAFGLYKLHMSCIIYD 211
Query 132 DLVNTDTVIERIEEIGL 148
D VNT+ +IE+IE I L
Sbjct 212 DFVNTNELIEKIENIDL 228
> xla:399468 eef1d; elongation factor-1 delta; K03232 elongation
factor 1-beta
Length=260
Score = 81.3 bits (199), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 42/107 (39%), Positives = 70/107 (65%), Gaps = 1/107 (0%)
Query 37 DLFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGNE 96
DLFG D E+ A +L + +Q +++ +K V+ KS ++++VKP D ETD+ ++
Sbjct 137 DLFGSDDEEEDAESARLREERLKQYAEKKSKKPGVIAKSSILLDVKPWDDETDMAKLEEC 196
Query 97 IKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIERI 143
++ +Q++GL WG + K VP+ +G+ KLQ+QCV+ DD V TD + E I
Sbjct 197 VRTVQMDGLVWG-SSKLVPVGYGIKKLQIQCVVEDDKVGTDILEEEI 242
> hsa:1936 EEF1D, EF-1D, EF1D, FLJ20897, FP1047; eukaryotic translation
elongation factor 1 delta (guanine nucleotide exchange
protein); K03232 elongation factor 1-beta
Length=647
Score = 81.3 bits (199), Expect = 1e-15, Method: Composition-based stats.
Identities = 42/106 (39%), Positives = 68/106 (64%), Gaps = 1/106 (0%)
Query 38 LFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGNEI 97
LFG D E+ QL + +Q +++ +K +V KS ++++VKP D ETD+ ++ +
Sbjct 525 LFGSDNEEEDKEAAQLREERLRQYAEKKAKKPALVAKSSILLDVKPWDDETDMAQLEACV 584
Query 98 KQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIERI 143
+ IQ++GL WG + K VP+ +G+ KLQ+QCV+ DD V TD + E I
Sbjct 585 RSIQLDGLVWGAS-KLVPVGYGIRKLQIQCVVEDDKVGTDLLEEEI 629
> xla:397892 eef1d; eukaryotic translation elongation factor 1
delta (guanine nucleotide exchange protein)
Length=265
Score = 81.3 bits (199), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/107 (38%), Positives = 70/107 (65%), Gaps = 1/107 (0%)
Query 37 DLFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGNE 96
DLFG D E+ A ++ + +Q +++ +K V+ KS ++++VKP D ETD+ ++
Sbjct 142 DLFGSDNEEEDAEAARIREERLKQYAEKKSKKPGVIAKSSILLDVKPWDDETDMAKLEEC 201
Query 97 IKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIERI 143
++ +Q++GL WG + K VP+ +G+ KLQ+QCV+ DD V TD + E I
Sbjct 202 VRTVQMDGLVWG-SSKLVPVGYGIKKLQIQCVVEDDKVGTDILEEEI 247
> mmu:55949 Eef1b2, 2810017J07Rik, Eef1b; eukaryotic translation
elongation factor 1 beta 2; K03232 elongation factor 1-beta
Length=225
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 43/106 (40%), Positives = 69/106 (65%), Gaps = 1/106 (0%)
Query 38 LFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGNEI 97
LFG D E+ + K+L + Q + ++ +K VV KS ++++VKP D ETD+ ++ +
Sbjct 103 LFGSDDEEESEEAKKLREERLAQYESKKAKKPAVVAKSSILLDVKPWDDETDMTKLEECV 162
Query 98 KQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIERI 143
+ IQ +GL WG + K VP+ +G+ KLQ+QCV+ DD V TD + E+I
Sbjct 163 RSIQADGLVWG-SSKLVPVGYGIKKLQIQCVVEDDKVGTDMLEEQI 207
> dre:100332692 elongation factor-1, delta-like
Length=450
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 40/106 (37%), Positives = 71/106 (66%), Gaps = 3/106 (2%)
Query 38 LFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGNEI 97
LFG D ED A +++ A + ++ Q++ +K ++ KS ++++VKP D ETD++++ +
Sbjct 330 LFGSDEEDEEA--ERIKAERVKEYSQRKAKKPALIAKSSILLDVKPWDDETDMSKLEECV 387
Query 98 KQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIERI 143
+ +Q++GL WG + K VP+ +G+ KLQ+ CV+ DD V TD + E I
Sbjct 388 RSVQMDGLLWGAS-KLVPVGYGIKKLQINCVVEDDKVGTDILEEEI 432
> dre:565501 eef1da, MGC111981, MGC55300, eef1d, zgc:66406; elongation
factor-1, delta, a
Length=163
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 40/106 (37%), Positives = 71/106 (66%), Gaps = 3/106 (2%)
Query 38 LFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGNEI 97
LFG D ED A +++ A + ++ Q++ +K ++ KS ++++VKP D ETD++++ +
Sbjct 43 LFGSDEEDEEA--ERIKAERVKEYSQRKAKKPALIAKSSILLDVKPWDDETDMSKLEECV 100
Query 98 KQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIERI 143
+ +Q++GL WG + K VP+ +G+ KLQ+ CV+ DD V TD + E I
Sbjct 101 RSVQMDGLLWGAS-KLVPVGYGIKKLQINCVVEDDKVGTDILEEEI 145
> dre:100331034 eef1d; elongation factor-1, delta-like
Length=439
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 40/109 (36%), Positives = 72/109 (66%), Gaps = 3/109 (2%)
Query 38 LFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGNEI 97
LFG D ED A +++ A + ++ Q++ +K ++ KS ++++VKP D ETD++++ +
Sbjct 319 LFGSDEEDEEA--ERIKAERVKEYSQRKAKKPALIAKSSILLDVKPWDDETDMSKLEECV 376
Query 98 KQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIERIEEI 146
+ +Q++GL WG + K VP+ +G+ KLQ+ CV+ DD V TD + E I +
Sbjct 377 RSVQMDGLLWGAS-KLVPVGYGIKKLQINCVVEDDKVGTDILEEEITQF 424
> hsa:1933 EEF1B2, EEF1B, EEF1B1, EF1B; eukaryotic translation
elongation factor 1 beta 2; K03232 elongation factor 1-beta
Length=225
Score = 79.0 bits (193), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 42/106 (39%), Positives = 69/106 (65%), Gaps = 1/106 (0%)
Query 38 LFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGNEI 97
LFG D E+ + K+L + Q + ++ +K +V KS ++++VKP D ETD+ ++ +
Sbjct 103 LFGSDDEEESEEAKRLREERLAQYESKKAKKPALVAKSSILLDVKPWDDETDMAKLEECV 162
Query 98 KQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIERI 143
+ IQ +GL WG + K VP+ +G+ KLQ+QCV+ DD V TD + E+I
Sbjct 163 RSIQADGLVWG-SSKLVPVGYGIKKLQIQCVVEDDKVGTDMLEEQI 207
> ath:AT1G30230 elongation factor 1-beta / EF-1-beta; K03232 elongation
factor 1-beta
Length=260
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/69 (50%), Positives = 51/69 (73%), Gaps = 1/69 (1%)
Query 73 NKSMLVIEVKPNDAETDLNEIGNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDD 132
KS ++I++KP D ETD+ ++ +K IQ+EGL WG + K VP+ +G+ KLQ+ C IVDD
Sbjct 141 GKSSVLIDIKPWDDETDMKKLEEAVKSIQMEGLFWGAS-KLVPVGYGIKKLQILCTIVDD 199
Query 133 LVNTDTVIE 141
LV+ DT+IE
Sbjct 200 LVSIDTMIE 208
> ath:AT5G12110 elongation factor 1B alpha-subunit 1 (eEF1Balpha1);
K03232 elongation factor 1-beta
Length=228
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 47/128 (36%), Positives = 78/128 (60%), Gaps = 7/128 (5%)
Query 14 PCYYYIHSQKESGKHATDDSDDIDLFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVN 73
P + H+++ + DD DDIDLF D+ ED + AA++++ ++ +K
Sbjct 85 PSEAHPHTEEPAADGDGDDDDDIDLFADETED------EKKAAEEREAAKKDTKKTKESG 138
Query 74 KSMLVIEVKPNDAETDLNEIGNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDL 133
KS +++EVKP D ETD+ ++ ++ +Q+ GLTWG + K VP+ +G+ KL + IVDDL
Sbjct 139 KSSVLLEVKPWDDETDMKKLEEAVRSVQMPGLTWGAS-KLVPVGYGIKKLTIMMTIVDDL 197
Query 134 VNTDTVIE 141
V+ D +IE
Sbjct 198 VSVDNLIE 205
> ath:AT2G18110 elongation factor 1-beta, putative / EF-1-beta,
putative; K03232 elongation factor 1-beta
Length=231
Score = 72.0 bits (175), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 32/69 (46%), Positives = 50/69 (72%), Gaps = 1/69 (1%)
Query 73 NKSMLVIEVKPNDAETDLNEIGNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDD 132
KS +++++KP D ETD+ ++ ++ IQ+EGL WG + K VP+ +G+ KL + C IVDD
Sbjct 141 GKSSVLMDIKPWDDETDMKKLEEAVRSIQMEGLFWGAS-KLVPVGYGIKKLHIMCTIVDD 199
Query 133 LVNTDTVIE 141
LV+ DT+IE
Sbjct 200 LVSIDTMIE 208
> sce:YAL003W EFB1, TEF5; Efb1p; K03232 elongation factor 1-beta
Length=206
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 33/71 (46%), Positives = 51/71 (71%), Gaps = 1/71 (1%)
Query 75 SMLVIEVKPNDAETDLNEIGNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLV 134
S++ ++VKP D ET+L E+ +K I++EGLTWG +Q +P+ FG+ KLQ+ CV+ DD V
Sbjct 121 SIVTLDVKPWDDETNLEEMVANVKAIEMEGLTWGAHQ-FIPIGFGIKKLQINCVVEDDKV 179
Query 135 NTDTVIERIEE 145
+ D + + IEE
Sbjct 180 SLDDLQQSIEE 190
> cel:F54H12.6 hypothetical protein; K03232 elongation factor
1-beta
Length=213
Score = 66.6 bits (161), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 38/108 (35%), Positives = 65/108 (60%), Gaps = 1/108 (0%)
Query 36 IDLFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGN 95
DLFG D E+ A ++ + +++ +K + KS ++++VKP D ETDL E+
Sbjct 88 FDLFGSDDEEEDAEKAKIVEERLAAYAEKKAKKAGPIAKSSVILDVKPWDDETDLGEMEK 147
Query 96 EIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIERI 143
++ I+++GL WG K +P+ +G+ KLQ+ VI D V+ D +IE+I
Sbjct 148 LVRSIEMDGLVWG-GAKLIPIGYGIKKLQIITVIEDLKVSVDDLIEKI 194
> cel:Y41E3.10 hypothetical protein; K03232 elongation factor
1-beta
Length=263
Score = 66.2 bits (160), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 39/110 (35%), Positives = 65/110 (59%), Gaps = 1/110 (0%)
Query 34 DDIDLFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEI 93
DD DLFG + E+ K++ + ++ K + KS ++++VKP D ETDL E+
Sbjct 136 DDFDLFGSEDEEEDEEKKKVVEERLAAYAAKKATKAGPIAKSSVILDVKPWDDETDLGEM 195
Query 94 GNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIERI 143
++ I+++GL WG K +P+ +G+ KLQ+ VI D V+ D +IE+I
Sbjct 196 EKLVRSIEMDGLVWG-GAKLIPIGYGIKKLQIITVIEDLKVSVDDLIEKI 244
> dre:560745 eef1db, MGC114052, eef1d, im:7233939, si:dkey-235d18.4,
wu:fa14f09, zgc:114052; elongation factor-1, delta, b;
K03232 elongation factor 1-beta
Length=274
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 49/69 (71%), Gaps = 1/69 (1%)
Query 75 SMLVIEVKPNDAETDLNEIGNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLV 134
S ++++VKP D ETD++++ ++ +Q++GL WG + K +P+ +G+ KLQ+ CV+ DD V
Sbjct 189 SSILLDVKPWDDETDMSKLEECVRSVQMDGLLWGAS-KLMPVGYGIKKLQINCVVEDDKV 247
Query 135 NTDTVIERI 143
TD + E I
Sbjct 248 GTDFLEEEI 256
> dre:335370 eef1b2, MGC86802, fj06d02, wu:fj06d02, zgc:56277,
zgc:86802; eukaryotic translation elongation factor 1 beta
2; K03232 elongation factor 1-beta
Length=225
Score = 61.6 bits (148), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 29/69 (42%), Positives = 49/69 (71%), Gaps = 1/69 (1%)
Query 75 SMLVIEVKPNDAETDLNEIGNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLV 134
S ++++VKP D ETD+ ++ ++ IQ++GL WG++ K +P+ +G+ KLQ+ CV+ DD V
Sbjct 140 SSILLDVKPWDDETDMAKLEECVRSIQLDGLVWGQS-KLLPVGYGIKKLQIACVVEDDKV 198
Query 135 NTDTVIERI 143
TD + E I
Sbjct 199 GTDQLEELI 207
> ath:AT5G19510 elongation factor 1B alpha-subunit 2 (eEF1Balpha2);
K03232 elongation factor 1-beta
Length=224
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 38/106 (35%), Positives = 66/106 (62%), Gaps = 7/106 (6%)
Query 36 IDLFGDDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGN 95
+DLFGD+ E+ A ++ AAK+ ++ + KS ++++VKP D ETD+ ++
Sbjct 103 MDLFGDETEEEKKAAEEREAAKKDTKKPKES------GKSSVLMDVKPWDDETDMKKLEE 156
Query 96 EIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIE 141
++ +++ GL WG + K VP+ +G+ KL + IVDDLV+ D +IE
Sbjct 157 AVRGVEMPGLFWGAS-KLVPVGYGIKKLTIMFTIVDDLVSPDNLIE 201
> tgo:TGME49_027830 mitochondrial import inner membrane translocase
TIM44, putative
Length=598
Score = 31.6 bits (70), Expect = 0.92, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 28/49 (57%), Gaps = 8/49 (16%)
Query 41 DDAEDSAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETD 89
DDAE AA +Q A K+ ++Q+ RK++Q + +P+D+E D
Sbjct 315 DDAEKKAAKWRQDMAVKRFKEQEARKREQELAQ--------RPSDSEAD 355
> mmu:28006 D6Wsu116e, A130095H06, C530005J20Rik, Fam21; DNA segment,
Chr 6, Wayne State University 116, expressed
Length=1334
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 5/49 (10%)
Query 36 IDLFGDDAED-----SAAALKQLSAAKQQQQQQQRKQKQPVVNKSMLVI 79
+ LFGD+ E+ SAAA KQ S+ + Q Q++ + +QP S L+
Sbjct 564 VSLFGDEDEEDSLFGSAAAKKQTSSLQPQSQEKAKPSEQPSKKTSALLF 612
> xla:398218 dazap1; DAZ associated protein 1; K14411 RNA-binding
protein Musashi
Length=360
Score = 30.8 bits (68), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 91 NEIGNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDD 132
N+ G+EI ++ + GL W Q+ + F Y V CVI+ D
Sbjct 3 NQGGDEIGKLFVGGLDWSTTQETLRSYFSQYGEVVDCVIMKD 44
> dre:561459 dazap1; DAZ associated protein 1; K14411 RNA-binding
protein Musashi
Length=418
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 24/46 (52%), Gaps = 0/46 (0%)
Query 87 ETDLNEIGNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDD 132
E + N G+EI ++ + GL W Q+ + F Y V CVI+ D
Sbjct 12 EMNSNLAGDEIGKLFVGGLDWSTTQETLRNYFSQYGEVVDCVIMKD 57
> hsa:29920 PYCR2, FLJ54750, P5CR2; pyrroline-5-carboxylate reductase
family, member 2 (EC:1.5.1.2); K00286 pyrroline-5-carboxylate
reductase [EC:1.5.1.2]
Length=320
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 25/97 (25%), Positives = 46/97 (47%), Gaps = 11/97 (11%)
Query 50 LKQLSAAKQQQQQQQRKQKQPVVNKSMLVIEVKPNDAETDLNEIGNEIKQIQI-----EG 104
L +SA ++ R K+ V + +L + VKP+ L+EIG +++ I G
Sbjct 39 LPTVSALRKMGVNLTRSNKETVKHSDVLFLAVKPHIIPFILDEIGADVQARHIVVSCAAG 98
Query 105 LTWGENQKKVPLAFGLYKLQVQCVIVDDLVNTDTVIE 141
+T +KK+ +AF ++C + NT V++
Sbjct 99 VTISSVEKKL-MAFQPAPKVIRC-----MTNTPVVVQ 129
> dre:568014 mKIAA0732 protein-like; K11124 protein SMG6 [EC:3.1.-.-]
Length=1546
Score = 28.9 bits (63), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 35/114 (30%), Positives = 54/114 (47%), Gaps = 16/114 (14%)
Query 24 ESGKHATDDSDDIDLFGDDAEDSAAALKQLS---AAKQQQQQQQRKQKQPVV-NKSMLVI 79
ES A++ D ++ GD +ED L+ A K QQQ++R + Q V+ L I
Sbjct 1307 ESSLSASEGEIDGEMEGDGSEDDIRELRARRHALAHKLAQQQKRRDKIQAVLQTGGQLEI 1366
Query 80 EVKPNDAETDLNEIGNEIKQIQIEGLTWGENQKKVPLAFGLYKLQVQCVIVDDL 133
EV+P D N + +EGL +K+ LA G Y L V +++ +L
Sbjct 1367 EVRPFYLVPDTNGFID-----HLEGL------RKL-LACGTYILVVPLIVITEL 1408
> pfa:PFC1011c conserved Plasmodium membrane protein, unknown
function
Length=729
Score = 28.5 bits (62), Expect = 7.9, Method: Composition-based stats.
Identities = 15/68 (22%), Positives = 37/68 (54%), Gaps = 4/68 (5%)
Query 18 YIHSQKESGKHATDDSDDIDLFGDDAED-SAAALKQLSAAKQQQQQQQRK---QKQPVVN 73
YI+ + ++ S DID+ DD ++ ++ + + + A ++Q+Q K +K +N
Sbjct 321 YIYILSSDSEKFSESSTDIDVLEDDLDNFKSSQIDEKNIASHMKEQKQNKYEEKKDDAIN 380
Query 74 KSMLVIEV 81
K++ + ++
Sbjct 381 KNVWIFKI 388
Lambda K H
0.311 0.130 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3134054208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40