bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2382_orf1
Length=115
Score E
Sequences producing significant alignments: (Bits) Value
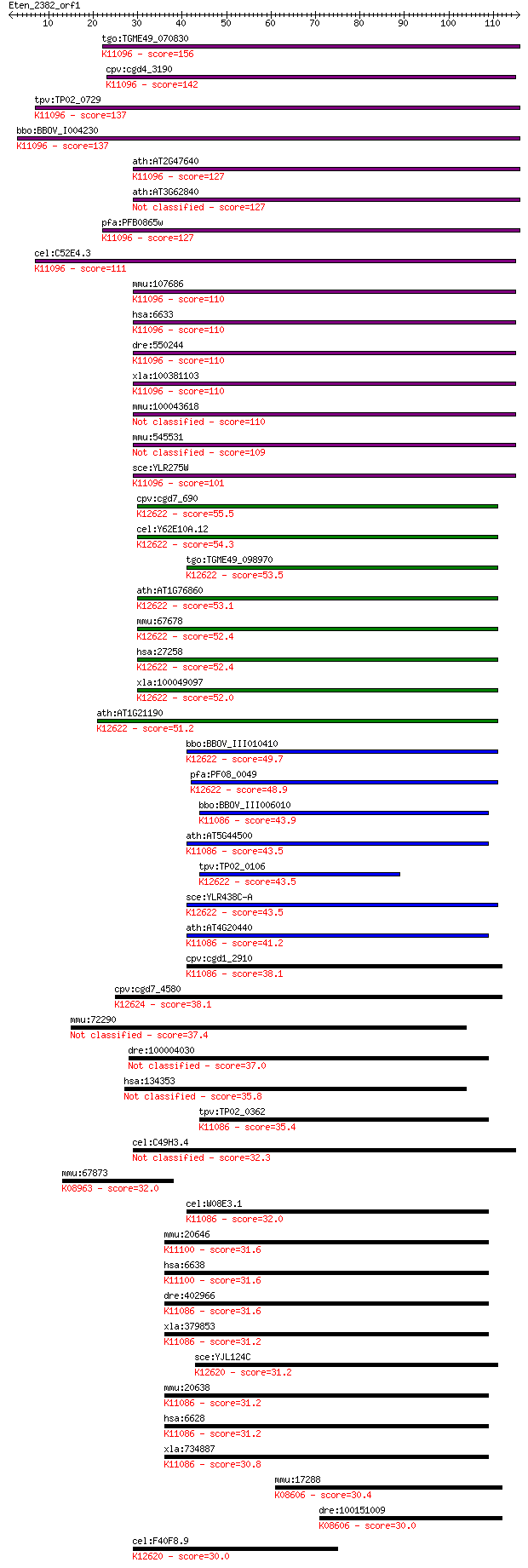
tgo:TGME49_070830 small nuclear ribonucleoprotein Sm D2, putat... 156 2e-38
cpv:cgd4_3190 small nuclear ribonucleoprotein ; K11096 small n... 142 2e-34
tpv:TP02_0729 small nuclear ribonucleoprotein D2; K11096 small... 137 6e-33
bbo:BBOV_I004230 19.m02306; small nuclear ribonucleoprotein; K... 137 7e-33
ath:AT2G47640 small nuclear ribonucleoprotein D2, putative / s... 127 7e-30
ath:AT3G62840 hypothetical protein 127 8e-30
pfa:PFB0865w small nuclear ribonucleoprotein, putative; K11096... 127 1e-29
cel:C52E4.3 snr-4; Small Nuclear Ribonucleoprotein family memb... 111 7e-25
mmu:107686 Snrpd2, 1810009A06Rik, MGC62953, SMD2; small nuclea... 110 8e-25
hsa:6633 SNRPD2, SMD2, SNRPD1, Sm-D2; small nuclear ribonucleo... 110 8e-25
dre:550244 snrpd2, im:6908977, si:dkey-113g17.2, zgc:110732; s... 110 8e-25
xla:100381103 snrpd2, sm-d2, smd2, snrpd1; small nuclear ribon... 110 9e-25
mmu:100043618 Gm10120; small nuclear ribonucleoprotein D2 pseu... 110 1e-24
mmu:545531 Gm5848, EG545531; predicted pseudogene 5848 109 2e-24
sce:YLR275W SMD2; Sm D2; K11096 small nuclear ribonucleoprotei... 101 7e-22
cpv:cgd7_690 small nuclear ribonucleoprotein ; K12622 U6 snRNA... 55.5 5e-08
cel:Y62E10A.12 lsm-3; LSM Sm-like protein family member (lsm-3... 54.3 1e-07
tgo:TGME49_098970 LSM domain-containing protein ; K12622 U6 sn... 53.5 2e-07
ath:AT1G76860 small nuclear ribonucleoprotein, putative / snRN... 53.1 2e-07
mmu:67678 Lsm3, 1010001J12Rik, 2610005D18Rik, 6030401D18Rik, S... 52.4 4e-07
hsa:27258 LSM3, SMX4, USS2, YLR438C; LSM3 homolog, U6 small nu... 52.4 4e-07
xla:100049097 lsm3; LSM3 homolog, U6 small nuclear RNA associa... 52.0 4e-07
ath:AT1G21190 small nuclear ribonucleoprotein, putative / snRN... 51.2 7e-07
bbo:BBOV_III010410 17.m07899; LSM domain containing protein; K... 49.7 3e-06
pfa:PF08_0049 lsm3 homologue, putative; K12622 U6 snRNA-associ... 48.9 4e-06
bbo:BBOV_III006010 17.m07530; Sm domain containing protein; K1... 43.9 1e-04
ath:AT5G44500 small nuclear ribonucleoprotein associated prote... 43.5 2e-04
tpv:TP02_0106 hypothetical protein; K12622 U6 snRNA-associated... 43.5 2e-04
sce:YLR438C-A LSM3, SMX4, USS2; Lsm (Like Sm) protein; part of... 43.5 2e-04
ath:AT4G20440 smB; smB (small nuclear ribonucleoprotein associ... 41.2 7e-04
cpv:cgd1_2910 hypothetical protein ; K11086 small nuclear ribo... 38.1 0.006
cpv:cgd7_4580 U6 snRNA-associated Sm-like protein LSm5. SM dom... 38.1 0.006
mmu:72290 Lsm11, 2210404M20Rik, AV079530; U7 snRNP-specific Sm... 37.4 0.013
dre:100004030 lsm11, im:7149708; LSM11, U7 small nuclear RNA a... 37.0 0.016
hsa:134353 LSM11, FLJ38273; LSM11, U7 small nuclear RNA associ... 35.8 0.038
tpv:TP02_0362 small nuclear ribonucleoprotein B; K11086 small ... 35.4 0.049
cel:C49H3.4 hypothetical protein 32.3 0.36
mmu:67873 Mri1, 2410018C20Rik; methylthioribose-1-phosphate is... 32.0 0.45
cel:W08E3.1 snr-2; Small Nuclear Ribonucleoprotein family memb... 32.0 0.56
mmu:20646 Snrpn, 2410045I01Rik, HCERN3, MGC18604, MGC30325, Pe... 31.6 0.61
hsa:6638 SNRPN, DKFZp686C0927, DKFZp686M12165, DKFZp761I1912, ... 31.6 0.61
dre:402966 snrpb, MGC77315, Snrpn, zgc:77315; small nuclear ri... 31.6 0.64
xla:379853 snrpn, MGC52854; small nuclear ribonucleoparticle-a... 31.2 0.74
sce:YJL124C LSM1, SPB8; Lsm (Like Sm) protein; forms heterohep... 31.2 0.76
mmu:20638 Snrpb, AL024368, AU018828, SM-B, SM11, SMB, SNRNP-B;... 31.2 0.95
hsa:6628 SNRPB, COD, SNRPB1, Sm-B/B', SmB/B', SmB/SmB', snRNP-... 31.2 0.95
xla:734887 snrpb, MGC130840, cod, sm-b/b', smb/b', smb/smb', s... 30.8 1.1
mmu:17288 Mep1b, MGC159330, Mep-1b; meprin 1 beta (EC:3.4.24.6... 30.4 1.5
dre:100151009 mep1b, MGC153272, si:ch211-191a24.6, zgc:153272;... 30.0 1.7
cel:F40F8.9 lsm-1; LSM Sm-like protein family member (lsm-1); ... 30.0 1.9
> tgo:TGME49_070830 small nuclear ribonucleoprotein Sm D2, putative
; K11096 small nuclear ribonucleoprotein D2
Length=104
Score = 156 bits (394), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 73/94 (77%), Positives = 85/94 (90%), Gaps = 0/94 (0%)
Query 22 NRDNPEGGPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTR 81
NRD+P GPL++L+ V DNSQVL+NCRNNRK+L RVKAFDRHCNL+LTDVREMW+E ++
Sbjct 11 NRDHPPEGPLSVLAASVKDNSQVLINCRNNRKVLGRVKAFDRHCNLVLTDVREMWTETSK 70
Query 82 GGGKKKQKVVNKDRFISKLFLRGDAVILILKNPK 115
GGGKKK + VNKDRFISKLFLRGDAVILIL+NPK
Sbjct 71 GGGKKKVRTVNKDRFISKLFLRGDAVILILRNPK 104
> cpv:cgd4_3190 small nuclear ribonucleoprotein ; K11096 small
nuclear ribonucleoprotein D2
Length=106
Score = 142 bits (359), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 64/92 (69%), Positives = 77/92 (83%), Gaps = 0/92 (0%)
Query 23 RDNPEGGPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRG 82
+DNP GPL+LL+ CV N+Q+LVNCRNNRK+L RVKAFDRHCNLLLTD RE+W+E +
Sbjct 14 KDNPSEGPLSLLTECVKTNTQILVNCRNNRKILGRVKAFDRHCNLLLTDAREIWTESMKS 73
Query 83 GGKKKQKVVNKDRFISKLFLRGDAVILILKNP 114
G K K +NKDRFISKLF+RGD+VILILK+P
Sbjct 74 GKKGSAKYINKDRFISKLFVRGDSVILILKSP 105
> tpv:TP02_0729 small nuclear ribonucleoprotein D2; K11096 small
nuclear ribonucleoprotein D2
Length=106
Score = 137 bits (346), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 66/109 (60%), Positives = 84/109 (77%), Gaps = 3/109 (2%)
Query 7 MAAAADGDKAAPHLVNRDNPEGGPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCN 66
M+ D L N+DNP GPL++L CV +N QVL+NCRNNRK+LARVKAFDRHCN
Sbjct 1 MSTKVKQDPTPMELDNKDNP-AGPLSILEECVRENCQVLINCRNNRKILARVKAFDRHCN 59
Query 67 LLLTDVREMWSEVTRGGGKKKQKVVNKDRFISKLFLRGDAVILILKNPK 115
++LT+VREMW+ RG G K+K+ KDRF+S+LFLRGD+VI++LKNPK
Sbjct 60 MILTNVREMWT--VRGKGTGKKKLETKDRFLSRLFLRGDSVIVVLKNPK 106
> bbo:BBOV_I004230 19.m02306; small nuclear ribonucleoprotein;
K11096 small nuclear ribonucleoprotein D2
Length=162
Score = 137 bits (346), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 68/113 (60%), Positives = 85/113 (75%), Gaps = 11/113 (9%)
Query 3 SKGIMAAAADGDKAAPHLVNRDNPEGGPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFD 62
S GI + + DK P GPL++L CV DNSQVL+NCR+NRKLL RVKAFD
Sbjct 61 SIGIRSMSTTEDKDCP---------SGPLSVLEACVRDNSQVLINCRSNRKLLGRVKAFD 111
Query 63 RHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDRFISKLFLRGDAVILILKNPK 115
RH N++LTDVREMW+EV+ GGGKK K +NKDR+IS+LFLRGD+V+++L NPK
Sbjct 112 RHFNMILTDVREMWTEVSGGGGKK--KYMNKDRYISRLFLRGDSVVVVLSNPK 162
> ath:AT2G47640 small nuclear ribonucleoprotein D2, putative /
snRNP core protein D2, putative / Sm protein D2, putative;
K11096 small nuclear ribonucleoprotein D2
Length=109
Score = 127 bits (320), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 59/88 (67%), Positives = 76/88 (86%), Gaps = 1/88 (1%)
Query 29 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGG-GKKK 87
GPL++L V +N+QVL+NCRNNRKLL RV+AFDRHCN++L +VREMW+EV + G GKKK
Sbjct 22 GPLSVLMMSVKNNTQVLINCRNNRKLLGRVRAFDRHCNMVLENVREMWTEVPKTGKGKKK 81
Query 88 QKVVNKDRFISKLFLRGDAVILILKNPK 115
VN+DRFISK+FLRGD+VI++L+NPK
Sbjct 82 ALPVNRDRFISKMFLRGDSVIIVLRNPK 109
> ath:AT3G62840 hypothetical protein
Length=108
Score = 127 bits (319), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 59/88 (67%), Positives = 76/88 (86%), Gaps = 1/88 (1%)
Query 29 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGG-GKKK 87
GPL++L V +N+QVL+NCRNNRKLL RV+AFDRHCN++L +VREMW+EV + G GKKK
Sbjct 21 GPLSVLMMSVKNNTQVLINCRNNRKLLGRVRAFDRHCNMVLENVREMWTEVPKTGKGKKK 80
Query 88 QKVVNKDRFISKLFLRGDAVILILKNPK 115
VN+DRFISK+FLRGD+VI++L+NPK
Sbjct 81 ALPVNRDRFISKMFLRGDSVIIVLRNPK 108
> pfa:PFB0865w small nuclear ribonucleoprotein, putative; K11096
small nuclear ribonucleoprotein D2
Length=101
Score = 127 bits (318), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 71/94 (75%), Positives = 81/94 (86%), Gaps = 2/94 (2%)
Query 22 NRDNPEGGPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTR 81
NRDNPE GPL LLS CV DN+QVL+NCRNNRK+L RVKAFDRHCNLLLT VRE+W EV +
Sbjct 10 NRDNPEDGPLGLLSECVKDNAQVLINCRNNRKILGRVKAFDRHCNLLLTGVREIWVEVVK 69
Query 82 GGGKKKQKVVNKDRFISKLFLRGDAVILILKNPK 115
KKK+K +NKDR+IS LFLRGD+VILIL+NPK
Sbjct 70 --DKKKKKKINKDRYISILFLRGDSVILILRNPK 101
> cel:C52E4.3 snr-4; Small Nuclear Ribonucleoprotein family member
(snr-4); K11096 small nuclear ribonucleoprotein D2
Length=118
Score = 111 bits (277), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 65/113 (57%), Positives = 85/113 (75%), Gaps = 5/113 (4%)
Query 7 MAAAAD--GDKAAPHLVNRDNPEG--GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFD 62
M+A A + A L +++ E GPL++L+ V +N QVL+NCRNN+KLL RVKAFD
Sbjct 1 MSAQAKPRSEMTAEELAAKEDEEFNVGPLSILTNSVKNNHQVLINCRNNKKLLGRVKAFD 60
Query 63 RHCNLLLTDVREMWSEV-TRGGGKKKQKVVNKDRFISKLFLRGDAVILILKNP 114
RHCN++L +V+EMW+EV G GKKK K V KDRFISK+FLRGD+VIL++KNP
Sbjct 61 RHCNMVLENVKEMWTEVPKTGKGKKKAKSVAKDRFISKMFLRGDSVILVVKNP 113
> mmu:107686 Snrpd2, 1810009A06Rik, MGC62953, SMD2; small nuclear
ribonucleoprotein D2; K11096 small nuclear ribonucleoprotein
D2
Length=118
Score = 110 bits (276), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 57/87 (65%), Positives = 76/87 (87%), Gaps = 1/87 (1%)
Query 29 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSE-VTRGGGKKK 87
GPL++L++ V +N+QVL+NCRNN+KLL RVKAFDRHCN++L +V+EMW+E G GKKK
Sbjct 27 GPLSVLTQSVKNNTQVLINCRNNKKLLGRVKAFDRHCNMVLENVKEMWTEVPKSGKGKKK 86
Query 88 QKVVNKDRFISKLFLRGDAVILILKNP 114
K VNKDR+ISK+FLRGD+VI++L+NP
Sbjct 87 SKPVNKDRYISKMFLRGDSVIVVLRNP 113
> hsa:6633 SNRPD2, SMD2, SNRPD1, Sm-D2; small nuclear ribonucleoprotein
D2 polypeptide 16.5kDa; K11096 small nuclear ribonucleoprotein
D2
Length=118
Score = 110 bits (276), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 57/87 (65%), Positives = 76/87 (87%), Gaps = 1/87 (1%)
Query 29 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSE-VTRGGGKKK 87
GPL++L++ V +N+QVL+NCRNN+KLL RVKAFDRHCN++L +V+EMW+E G GKKK
Sbjct 27 GPLSVLTQSVKNNTQVLINCRNNKKLLGRVKAFDRHCNMVLENVKEMWTEVPKSGKGKKK 86
Query 88 QKVVNKDRFISKLFLRGDAVILILKNP 114
K VNKDR+ISK+FLRGD+VI++L+NP
Sbjct 87 SKPVNKDRYISKMFLRGDSVIVVLRNP 113
> dre:550244 snrpd2, im:6908977, si:dkey-113g17.2, zgc:110732;
small nuclear ribonucleoprotein D2 polypeptide; K11096 small
nuclear ribonucleoprotein D2
Length=118
Score = 110 bits (276), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 57/87 (65%), Positives = 76/87 (87%), Gaps = 1/87 (1%)
Query 29 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSE-VTRGGGKKK 87
GPL++L++ V +N+QVL+NCRNN+KLL RVKAFDRHCN++L +V+EMW+E G GKKK
Sbjct 27 GPLSVLTQSVKNNTQVLINCRNNKKLLGRVKAFDRHCNMVLENVKEMWTEVPKSGKGKKK 86
Query 88 QKVVNKDRFISKLFLRGDAVILILKNP 114
K VNKDR+ISK+FLRGD+VI++L+NP
Sbjct 87 SKPVNKDRYISKMFLRGDSVIVVLRNP 113
> xla:100381103 snrpd2, sm-d2, smd2, snrpd1; small nuclear ribonucleoprotein
D2 polypeptide 16.5kDa; K11096 small nuclear
ribonucleoprotein D2
Length=118
Score = 110 bits (276), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 57/87 (65%), Positives = 76/87 (87%), Gaps = 1/87 (1%)
Query 29 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSE-VTRGGGKKK 87
GPL++L++ V +N+QVL+NCRNN+KLL RVKAFDRHCN++L +V+EMW+E G GKKK
Sbjct 27 GPLSVLTQSVKNNTQVLINCRNNKKLLGRVKAFDRHCNMVLENVKEMWTEVPKSGKGKKK 86
Query 88 QKVVNKDRFISKLFLRGDAVILILKNP 114
K VNKDR+ISK+FLRGD+VI++L+NP
Sbjct 87 SKPVNKDRYISKMFLRGDSVIVVLRNP 113
> mmu:100043618 Gm10120; small nuclear ribonucleoprotein D2 pseudogene
Length=118
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 57/87 (65%), Positives = 75/87 (86%), Gaps = 1/87 (1%)
Query 29 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSE-VTRGGGKKK 87
GPL++L++ V +N+QVL+NCRNN+KLL RVKAFDRHCN++L +V+EMW+E G GKKK
Sbjct 27 GPLSVLTQSVKNNTQVLINCRNNKKLLGRVKAFDRHCNMVLENVKEMWTEVPKSGKGKKK 86
Query 88 QKVVNKDRFISKLFLRGDAVILILKNP 114
K VNKDR+ISK+FLRGD VI++L+NP
Sbjct 87 SKPVNKDRYISKMFLRGDLVIVVLRNP 113
> mmu:545531 Gm5848, EG545531; predicted pseudogene 5848
Length=118
Score = 109 bits (272), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 53/87 (60%), Positives = 73/87 (83%), Gaps = 1/87 (1%)
Query 29 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQ 88
GPL++LS+ V +N+QVL+NCRN +KLL RVKAFDRHCN++L +V+EMW+EV + KK+
Sbjct 27 GPLSVLSQSVKNNTQVLINCRNKKKLLGRVKAFDRHCNMVLENVKEMWTEVPKSSKGKKK 86
Query 89 KV-VNKDRFISKLFLRGDAVILILKNP 114
VNKD +ISK+FLRGD+VI++L+NP
Sbjct 87 SKPVNKDCYISKIFLRGDSVIVVLRNP 113
> sce:YLR275W SMD2; Sm D2; K11096 small nuclear ribonucleoprotein
D2
Length=110
Score = 101 bits (251), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 44/86 (51%), Positives = 69/86 (80%), Gaps = 6/86 (6%)
Query 29 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQ 88
GP++L++ ++ + V+++ RNN K++ARVKAFDRHCN++L +V+E+W+E KK +
Sbjct 29 GPMSLINDAMVTRTPVIISLRNNHKIIARVKAFDRHCNMVLENVKELWTE------KKGK 82
Query 89 KVVNKDRFISKLFLRGDAVILILKNP 114
V+N++RFISKLFLRGD+VI++LK P
Sbjct 83 NVINRERFISKLFLRGDSVIVVLKTP 108
> cpv:cgd7_690 small nuclear ribonucleoprotein ; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=95
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 33/81 (40%), Positives = 45/81 (55%), Gaps = 2/81 (2%)
Query 30 PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQK 89
PL L+ R LD QV V CR NR+L + A+D H N++L +V E + E + +
Sbjct 7 PLDLI-RLSLD-EQVFVKCRGNRELKGTLYAYDPHMNMVLGNVEETYYEEESKSDTQNSE 64
Query 90 VVNKDRFISKLFLRGDAVILI 110
K R I LFLRGD +IL+
Sbjct 65 KKLKKRRIEMLFLRGDLIILV 85
> cel:Y62E10A.12 lsm-3; LSM Sm-like protein family member (lsm-3);
K12622 U6 snRNA-associated Sm-like protein LSm3
Length=102
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 34/83 (40%), Positives = 52/83 (62%), Gaps = 6/83 (7%)
Query 30 PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQK 89
PL LL R LD +V V RN+R+L R++AFD+H N++L++V E + TR + +
Sbjct 17 PLDLL-RLSLDE-RVYVKMRNDRELRGRLRAFDQHLNMVLSEVEETIT--TREVDEDTFE 72
Query 90 VVNKD--RFISKLFLRGDAVILI 110
+ K R + LF+RGD+VIL+
Sbjct 73 EIYKQTKRVVPMLFVRGDSVILV 95
> tgo:TGME49_098970 LSM domain-containing protein ; K12622 U6
snRNA-associated Sm-like protein LSm3
Length=96
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 43/70 (61%), Gaps = 0/70 (0%)
Query 41 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDRFISKL 100
+ +V + CR +R+++ ++ A+D H N++L DV E+ + VT ++ R + +
Sbjct 19 DERVTIKCRGDREVVGKLHAYDMHLNMVLGDVEEVATTVTSDPLTGDEQTKKTTRRLPLI 78
Query 101 FLRGDAVILI 110
FLRGDA+IL+
Sbjct 79 FLRGDAIILV 88
> ath:AT1G76860 small nuclear ribonucleoprotein, putative / snRNP,
putative / Sm protein, putative; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=98
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/81 (38%), Positives = 47/81 (58%), Gaps = 2/81 (2%)
Query 30 PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQK 89
PL L+ R LD ++ V R++R+L ++ AFD+H N++L DV E + V ++
Sbjct 12 PLDLI-RLSLDE-RIYVKLRSDRELRGKLHAFDQHLNMILGDVEETITTVEIDDETYEEI 69
Query 90 VVNKDRFISKLFLRGDAVILI 110
V R I LF+RGD VIL+
Sbjct 70 VRTTKRTIEFLFVRGDGVILV 90
> mmu:67678 Lsm3, 1010001J12Rik, 2610005D18Rik, 6030401D18Rik,
SMX4, USS2; LSM3 homolog, U6 small nuclear RNA associated (S.
cerevisiae); K12622 U6 snRNA-associated Sm-like protein LSm3
Length=102
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 47/81 (58%), Gaps = 2/81 (2%)
Query 30 PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQK 89
PL L+ R LD ++ V RN+R+L R+ A+D+H N++L DV E + + ++
Sbjct 17 PLDLI-RLSLD-ERIYVKMRNDRELRGRLHAYDQHLNMILGDVEETVTTIEIDEETYEEI 74
Query 90 VVNKDRFISKLFLRGDAVILI 110
+ R I LF+RGD V+L+
Sbjct 75 YKSTKRNIPMLFVRGDGVVLV 95
> hsa:27258 LSM3, SMX4, USS2, YLR438C; LSM3 homolog, U6 small
nuclear RNA associated (S. cerevisiae); K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=102
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 47/81 (58%), Gaps = 2/81 (2%)
Query 30 PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQK 89
PL L+ R LD ++ V RN+R+L R+ A+D+H N++L DV E + + ++
Sbjct 17 PLDLI-RLSLD-ERIYVKMRNDRELRGRLHAYDQHLNMILGDVEETVTTIEIDEETYEEI 74
Query 90 VVNKDRFISKLFLRGDAVILI 110
+ R I LF+RGD V+L+
Sbjct 75 YKSTKRNIPMLFVRGDGVVLV 95
> xla:100049097 lsm3; LSM3 homolog, U6 small nuclear RNA associated;
K12622 U6 snRNA-associated Sm-like protein LSm3
Length=102
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 47/81 (58%), Gaps = 2/81 (2%)
Query 30 PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQK 89
PL L+ R LD ++ V RN+R+L R+ A+D+H N++L DV E + + ++
Sbjct 17 PLDLI-RLSLD-ERIYVKMRNDRELRGRLNAYDQHLNMILGDVEETVTTIEIDEETYEEI 74
Query 90 VVNKDRFISKLFLRGDAVILI 110
+ R I LF+RGD V+L+
Sbjct 75 YKSTKRNIPMLFVRGDGVVLV 95
> ath:AT1G21190 small nuclear ribonucleoprotein, putative / snRNP,
putative / Sm protein, putative; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=97
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 48/90 (53%), Gaps = 2/90 (2%)
Query 21 VNRDNPEGGPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVT 80
V D PL L+ + ++ V R++R+L ++ AFD+H N++L DV E+ + +
Sbjct 3 VEEDATVREPLDLIRLSI--EERIYVKLRSDRELRGKLHAFDQHLNMILGDVEEVITTIE 60
Query 81 RGGGKKKQKVVNKDRFISKLFLRGDAVILI 110
++ V R + LF+RGD VIL+
Sbjct 61 IDDETYEEIVRTTKRTVPFLFVRGDGVILV 90
> bbo:BBOV_III010410 17.m07899; LSM domain containing protein;
K12622 U6 snRNA-associated Sm-like protein LSm3
Length=89
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 46/72 (63%), Gaps = 4/72 (5%)
Query 41 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWS--EVTRGGGKKKQKVVNKDRFIS 98
+ +V + C+ R+++ ++ A+D HCN+LL+D E ++ E+ ++ + +KD +
Sbjct 14 DERVYLKCKGGREIVGQLHAYDEHCNMLLSDAIETFTSVELDPTTNQEVTTITSKDSGV- 72
Query 99 KLFLRGDAVILI 110
+F+RGDA+IL+
Sbjct 73 -VFVRGDALILL 83
> pfa:PF08_0049 lsm3 homologue, putative; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=91
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 44/69 (63%), Gaps = 2/69 (2%)
Query 42 SQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDRFISKLF 101
++ + C+ +R+L+ ++ A+D H N++L++VRE + + ++ V +R + +F
Sbjct 20 EEIFLKCKGDRELIGKLDAYDNHLNMILSNVRETYKYSVKEND--EETVKKMERNLDMVF 77
Query 102 LRGDAVILI 110
+RGD++IL+
Sbjct 78 VRGDSIILV 86
> bbo:BBOV_III006010 17.m07530; Sm domain containing protein;
K11086 small nuclear ribonucleoprotein B and B'
Length=159
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 37/65 (56%), Gaps = 3/65 (4%)
Query 44 VLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDRFISKLFLR 103
V V ++ RK + AFD+H NL+L D E +T+G G KQKV K R + + LR
Sbjct 16 VRVAIKDGRKFVGTFIAFDKHMNLVLADCEEF--RITKGKGPDKQKVELK-RTLGFIMLR 72
Query 104 GDAVI 108
G+ ++
Sbjct 73 GENIV 77
> ath:AT5G44500 small nuclear ribonucleoprotein associated protein
B, putative / snRNP-B, putative / Sm protein B, putative;
K11086 small nuclear ribonucleoprotein B and B'
Length=254
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 36/68 (52%), Gaps = 0/68 (0%)
Query 41 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDRFISKL 100
N ++ V ++ R+L+ + AFDRH NL+L D E G KK + + R + +
Sbjct 14 NYRMRVTIQDGRQLIGKFMAFDRHMNLVLGDCEEFRKLPPAKGNKKTNEEREERRTLGLV 73
Query 101 FLRGDAVI 108
LRG+ VI
Sbjct 74 LLRGEEVI 81
> tpv:TP02_0106 hypothetical protein; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=84
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 44 VLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQ 88
+ + C+N R+L+ R++AFD HCN++L++V E + V K +
Sbjct 22 IYLKCKNGRELVGRLQAFDEHCNMVLSEVTETITTVELDNNSKNE 66
> sce:YLR438C-A LSM3, SMX4, USS2; Lsm (Like Sm) protein; part
of heteroheptameric complexes (Lsm2p-7p and either Lsm1p or
8p): cytoplasmic Lsm1p complex involved in mRNA decay; nuclear
Lsm8p complex part of U6 snRNP and possibly involved in processing
tRNA, snoRNA, and rRNA; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=89
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 38/70 (54%), Gaps = 6/70 (8%)
Query 41 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDRFISKL 100
+ +V + R R L+ ++AFD HCN++L+D E ++ +++ +R +
Sbjct 13 DERVYIKLRGARTLVGTLQAFDSHCNIVLSDAVETIYQL------NNEELSESERRCEMV 66
Query 101 FLRGDAVILI 110
F+RGD V LI
Sbjct 67 FIRGDTVTLI 76
> ath:AT4G20440 smB; smB (small nuclear ribonucleoprotein associated
protein B); K11086 small nuclear ribonucleoprotein B
and B'
Length=257
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 38/68 (55%), Gaps = 1/68 (1%)
Query 41 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDRFISKL 100
N ++ V ++ R+L+ + AFDRH NL+L D E + ++ GKK + R + +
Sbjct 14 NYRMRVTIQDGRQLVGKFMAFDRHMNLVLGDCEE-FRKLPPAKGKKINEEREDRRTLGLV 72
Query 101 FLRGDAVI 108
LRG+ VI
Sbjct 73 LLRGEEVI 80
> cpv:cgd1_2910 hypothetical protein ; K11086 small nuclear ribonucleoprotein
B and B'
Length=139
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 41/71 (57%), Gaps = 6/71 (8%)
Query 41 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDRFISKL 100
N ++ V +++R ++ + AFDRH NL+L+D +E + V +G K+ K R + +
Sbjct 6 NYRIRVTVQDDRVMVGNLMAFDRHMNLVLSDCQE-YRRVKKGEESKELK-----RSLGLI 59
Query 101 FLRGDAVILIL 111
LRG+ ++ +
Sbjct 60 MLRGENIVTFV 70
> cpv:cgd7_4580 U6 snRNA-associated Sm-like protein LSm5. SM domain
; K12624 U6 snRNA-associated Sm-like protein LSm5
Length=121
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 26/94 (27%), Positives = 50/94 (53%), Gaps = 10/94 (10%)
Query 25 NPEGG----PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMW---S 77
N +GG PL L+ +C+ +++ V + +++ ++ FD + N++L DV+E
Sbjct 20 NQKGGNIILPLALIDKCI--GNRIYVVMKGDKEFSGVLRGFDEYVNMVLDDVQEYGFKAD 77
Query 78 EVTRGGGKKKQKVVNKDRFISKLFLRGDAVILIL 111
E GG KK K V +R + + L G+ V +++
Sbjct 78 EEDISGGNKKLKRVMVNR-LETILLSGNNVAMLV 110
> mmu:72290 Lsm11, 2210404M20Rik, AV079530; U7 snRNP-specific
Sm-like protein LSM11
Length=361
Score = 37.4 bits (85), Expect = 0.013, Method: Composition-based stats.
Identities = 29/95 (30%), Positives = 44/95 (46%), Gaps = 6/95 (6%)
Query 15 KAAPHLVNRDNP--EGGPLTLLSRCVLDNSQVLVNCRNNRKL----LARVKAFDRHCNLL 68
K AP V P EG PL L RC+ + +V V+ R + L + AFD+ N+
Sbjct 139 KKAPRNVLTRMPLHEGSPLGELHRCIREGVKVNVHIRTFKGLRGVCTGFLVAFDKFWNMA 198
Query 69 LTDVREMWSEVTRGGGKKKQKVVNKDRFISKLFLR 103
LTDV E + + G ++ + R +L L+
Sbjct 199 LTDVDETYRKPVLGKAYERDSSLTLTRLFDRLKLQ 233
> dre:100004030 lsm11, im:7149708; LSM11, U7 small nuclear RNA
associated
Length=363
Score = 37.0 bits (84), Expect = 0.016, Method: Composition-based stats.
Identities = 27/85 (31%), Positives = 40/85 (47%), Gaps = 4/85 (4%)
Query 28 GGPLTLLSRCVLDNSQVLVNCRNNRKLLAR----VKAFDRHCNLLLTDVREMWSEVTRGG 83
G PL L+RCV + +V V+ R + L V AFD+ NL + DV E + E G
Sbjct 189 GSPLGQLNRCVQEKIRVKVHIRTFKGLRGVCSGFVVAFDKFWNLAMVDVDETYREPLLGQ 248
Query 84 GKKKQKVVNKDRFISKLFLRGDAVI 108
+K + R KL ++ A +
Sbjct 249 ALYHEKALTITRLFEKLKVQETAAL 273
> hsa:134353 LSM11, FLJ38273; LSM11, U7 small nuclear RNA associated
Length=360
Score = 35.8 bits (81), Expect = 0.038, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 39/81 (48%), Gaps = 4/81 (4%)
Query 27 EGGPLTLLSRCVLDNSQVLVNCRNNRKL----LARVKAFDRHCNLLLTDVREMWSEVTRG 82
EG PL L RC+ + +V V+ R + L + AFD+ N+ LTDV E + + G
Sbjct 152 EGSPLGELHRCIREGVKVNVHIRTFKGLRGVCTGFLVAFDKFWNMALTDVDETYRKPVLG 211
Query 83 GGKKKQKVVNKDRFISKLFLR 103
++ + R +L L+
Sbjct 212 KAYERDSSLTLTRLFDRLKLQ 232
> tpv:TP02_0362 small nuclear ribonucleoprotein B; K11086 small
nuclear ribonucleoprotein B and B'
Length=155
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 37/65 (56%), Gaps = 5/65 (7%)
Query 44 VLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDRFISKLFLR 103
V V ++NRK + + A+D++ NL+L+D E +T G K + +V R + + LR
Sbjct 16 VRVTLKDNRKFVGTLVAYDKYMNLVLSDCEEF--RMTLGKDKNRTEV---KRTLGFVLLR 70
Query 104 GDAVI 108
G+ ++
Sbjct 71 GENIV 75
> cel:C49H3.4 hypothetical protein
Length=223
Score = 32.3 bits (72), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 28/120 (23%), Positives = 48/120 (40%), Gaps = 38/120 (31%)
Query 29 GPLTLLSRCVLDNSQVLVNCRN----NRKLLARVKAFDRHCNLLLTDVREM--------- 75
GP+++++ V++ ++ V R+ +R FD H NL++ DV E
Sbjct 108 GPMSMIANAVINQKRIEVRVRSAIRIDRIFEGIPTTFDEHFNLMMKDVTETVLHGRKAQK 167
Query 76 -----------------WSEVTRGGGKKKQKV----VNKDRFISKLFLRGDAVILILKNP 114
W E GG KV + + RF F++GD+V+L+ P
Sbjct 168 EFTNRKEMQLLLPEFLRWKE----GGNWPMKVGTNRLIEHRFQRACFIKGDSVVLVRMLP 223
> mmu:67873 Mri1, 2410018C20Rik; methylthioribose-1-phosphate
isomerase homolog (S. cerevisiae) (EC:5.3.1.23); K08963 methylthioribose-1-phosphate
isomerase [EC:5.3.1.23]
Length=369
Score = 32.0 bits (71), Expect = 0.45, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 13 GDKAAPHLVNRDNPEGGPLTLLSRC 37
GD A HL+ + NP GG +T+L+ C
Sbjct 144 GDLGARHLLEQTNPRGGKVTVLTHC 168
> cel:W08E3.1 snr-2; Small Nuclear Ribonucleoprotein family member
(snr-2); K11086 small nuclear ribonucleoprotein B and B'
Length=160
Score = 32.0 bits (71), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 17/68 (25%), Positives = 35/68 (51%), Gaps = 3/68 (4%)
Query 41 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDRFISKL 100
N ++ + ++ R + KAFD+H N+LL + E ++ GKK + R + +
Sbjct 14 NYRMKIILQDGRTFIGFFKAFDKHMNILLAECEE-HRQIKPKAGKKTDG--EEKRILGLV 70
Query 101 FLRGDAVI 108
+RG+ ++
Sbjct 71 LVRGEHIV 78
> mmu:20646 Snrpn, 2410045I01Rik, HCERN3, MGC18604, MGC30325,
Peg4, SMN; small nuclear ribonucleoprotein N; K11100 small nuclear
ribonucleoprotein N
Length=240
Score = 31.6 bits (70), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 33/73 (45%), Gaps = 12/73 (16%)
Query 36 RCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDR 95
RC+L + ++ + KAFD+H NL+L D E + KQ + R
Sbjct 18 RCILQDGRIFIGT---------FKAFDKHMNLILCDCDEFRKIKPK---NAKQPEREEKR 65
Query 96 FISKLFLRGDAVI 108
+ + LRG+ ++
Sbjct 66 VLGLVLLRGENLV 78
> hsa:6638 SNRPN, DKFZp686C0927, DKFZp686M12165, DKFZp761I1912,
DKFZp762N022, FLJ33569, FLJ36996, FLJ39265, HCERN3, MGC29886,
PWCR, RT-LI, SM-D, SMN, SNRNP-N, SNURF-SNRPN; small nuclear
ribonucleoprotein polypeptide N; K11100 small nuclear ribonucleoprotein
N
Length=240
Score = 31.6 bits (70), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 33/73 (45%), Gaps = 12/73 (16%)
Query 36 RCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDR 95
RC+L + ++ + KAFD+H NL+L D E + KQ + R
Sbjct 18 RCILQDGRIFIGT---------FKAFDKHMNLILCDCDEFRKIKPK---NAKQPEREEKR 65
Query 96 FISKLFLRGDAVI 108
+ + LRG+ ++
Sbjct 66 VLGLVLLRGENLV 78
> dre:402966 snrpb, MGC77315, Snrpn, zgc:77315; small nuclear
ribonucleoprotein polypeptides B and B1; K11086 small nuclear
ribonucleoprotein B and B'
Length=239
Score = 31.6 bits (70), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 33/73 (45%), Gaps = 12/73 (16%)
Query 36 RCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDR 95
RC+L + ++ + KAFD+H NL+L D E + KQ + R
Sbjct 18 RCILQDGRIFIGT---------FKAFDKHMNLILCDCDEFRKIKPK---NSKQPEREEKR 65
Query 96 FISKLFLRGDAVI 108
+ + LRG+ ++
Sbjct 66 VLGLVLLRGENLV 78
> xla:379853 snrpn, MGC52854; small nuclear ribonucleoparticle-associated
protein (snRNP) mRNA, clone Sm51; K11086 small nuclear
ribonucleoprotein B and B'
Length=236
Score = 31.2 bits (69), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 33/73 (45%), Gaps = 12/73 (16%)
Query 36 RCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDR 95
RC+L + ++ + KAFD+H NL+L D E + KQ + R
Sbjct 18 RCILQDGRIFIGT---------FKAFDKHMNLILCDCDEFRKIKPK---NSKQPEREEKR 65
Query 96 FISKLFLRGDAVI 108
+ + LRG+ ++
Sbjct 66 VLGLVLLRGENLV 78
> sce:YJL124C LSM1, SPB8; Lsm (Like Sm) protein; forms heteroheptameric
complex (with Lsm2p, Lsm3p, Lsm4p, Lsm5p, Lsm6p, and
Lsm7p) involved in degradation of cytoplasmic mRNAs; K12620
U6 snRNA-associated Sm-like protein LSm1
Length=172
Score = 31.2 bits (69), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 35/68 (51%), Gaps = 8/68 (11%)
Query 43 QVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDRFISKLFL 102
++ V R+ R L ++ FD++ NL+L D V R ++ K +DR I +
Sbjct 53 KIFVLLRDGRMLFGVLRTFDQYANLILQDC------VERIYFSEENKYAEEDRGI--FMI 104
Query 103 RGDAVILI 110
RG+ V+++
Sbjct 105 RGENVVML 112
> mmu:20638 Snrpb, AL024368, AU018828, SM-B, SM11, SMB, SNRNP-B;
small nuclear ribonucleoprotein B; K11086 small nuclear ribonucleoprotein
B and B'
Length=231
Score = 31.2 bits (69), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 33/73 (45%), Gaps = 12/73 (16%)
Query 36 RCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDR 95
RC+L + ++ + KAFD+H NL+L D E + KQ + R
Sbjct 18 RCILQDGRIFIGT---------FKAFDKHMNLILCDCDEFRKIKPK---NSKQAEREEKR 65
Query 96 FISKLFLRGDAVI 108
+ + LRG+ ++
Sbjct 66 VLGLVLLRGENLV 78
> hsa:6628 SNRPB, COD, SNRPB1, Sm-B/B', SmB/B', SmB/SmB', snRNP-B;
small nuclear ribonucleoprotein polypeptides B and B1;
K11086 small nuclear ribonucleoprotein B and B'
Length=231
Score = 31.2 bits (69), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 33/73 (45%), Gaps = 12/73 (16%)
Query 36 RCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDR 95
RC+L + ++ + KAFD+H NL+L D E + KQ + R
Sbjct 18 RCILQDGRIFIGT---------FKAFDKHMNLILCDCDEFRKIKPK---NSKQAEREEKR 65
Query 96 FISKLFLRGDAVI 108
+ + LRG+ ++
Sbjct 66 VLGLVLLRGENLV 78
> xla:734887 snrpb, MGC130840, cod, sm-b/b', smb/b', smb/smb',
snrnp-b, snrpb1; small nuclear ribonucleoprotein polypeptides
B and B1; K11086 small nuclear ribonucleoprotein B and B'
Length=234
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 32/73 (43%), Gaps = 12/73 (16%)
Query 36 RCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDR 95
RC L + ++ + KAFD+H NL+L D E + KQ + R
Sbjct 18 RCTLQDGRIFIGT---------FKAFDKHMNLILCDCDEFRKIKPK---NSKQPEREEKR 65
Query 96 FISKLFLRGDAVI 108
+ + LRG+ ++
Sbjct 66 VLGLVLLRGENLV 78
> mmu:17288 Mep1b, MGC159330, Mep-1b; meprin 1 beta (EC:3.4.24.63);
K08606 meprin A, beta [EC:3.4.24.18]
Length=704
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 14/51 (27%), Positives = 29/51 (56%), Gaps = 0/51 (0%)
Query 61 FDRHCNLLLTDVREMWSEVTRGGGKKKQKVVNKDRFISKLFLRGDAVILIL 111
+DR + +TDV ++ +RG G + ++R S+ F++GD + ++L
Sbjct 534 WDRPSKVGVTDVFPNGTQFSRGIGYGTTVFITRERLKSREFIKGDDIYILL 584
> dre:100151009 mep1b, MGC153272, si:ch211-191a24.6, zgc:153272;
meprin A, beta (EC:3.4.24.18); K08606 meprin A, beta [EC:3.4.24.18]
Length=677
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 8/49 (16%)
Query 71 DVREMWSEVT--------RGGGKKKQKVVNKDRFISKLFLRGDAVILIL 111
D R++ S VT RG G + DR S+ F++GD VI +L
Sbjct 548 DPRKVGSRVTDTDGSTFYRGSGYGTSSFITHDRLKSRSFIKGDDVIFLL 596
> cel:F40F8.9 lsm-1; LSM Sm-like protein family member (lsm-1);
K12620 U6 snRNA-associated Sm-like protein LSm1
Length=125
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 28/46 (60%), Gaps = 3/46 (6%)
Query 29 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVRE 74
G ++L + + ++LV R+ RKL+ +++ D+ NL+L DV E
Sbjct 10 GAISLFEQL---DKKLLVVLRDGRKLIGFLRSIDQFANLILEDVVE 52
Lambda K H
0.321 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2042676840
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40