bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2395_orf1
Length=179
Score E
Sequences producing significant alignments: (Bits) Value
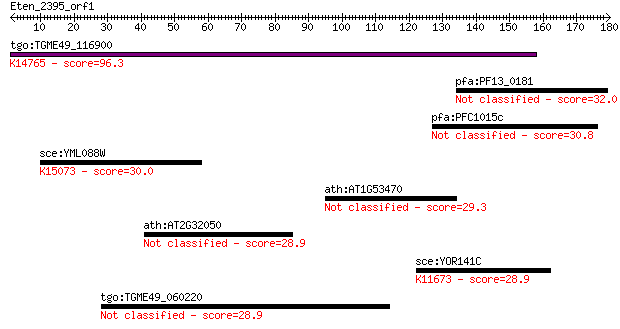
tgo:TGME49_116900 hypothetical protein ; K14765 U3 small nucle... 96.3 5e-20
pfa:PF13_0181 conserved Plasmodium protein, unknown function 32.0 1.0
pfa:PFC1015c conserved Plasmodium protein, unknown function 30.8 2.5
sce:YML088W UFO1; F-box receptor protein, subunit of the Skp1-... 30.0 5.0
ath:AT1G53470 MSL4; MSL4 (MECHANOSENSITIVE CHANNEL OF SMALL CO... 29.3 7.6
ath:AT2G32050 cell cycle control protein-related 28.9 8.6
sce:YOR141C ARP8; Arp8p; K11673 actin-related protein 8 28.9
tgo:TGME49_060220 folate/methotrexate transporter, putative 28.9 9.3
> tgo:TGME49_116900 hypothetical protein ; K14765 U3 small nucleolar
ribonucleoprotein protein LCP5
Length=800
Score = 96.3 bits (238), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 64/168 (38%), Positives = 103/168 (61%), Gaps = 20/168 (11%)
Query 1 RREMVRAVKEMTGDAPEEVGIERWL---------AAKAHKEAAREAGLVGDDEDMQLMRR 51
R E+VRAV+E GDAPEEVG+E+W+ AA K+ AREA E+ ++R
Sbjct 479 RSELVRAVREEVGDAPEEVGLEQWIQVQQSRLGSAASIAKQRAREAF-----EEENMVRL 533
Query 52 SLSKKERKVQAAQRALFERRAALSVGSTLEDLSIFCEEELGGVGEDDDGPFDLQSRGKG- 110
S+SKK+RK + + L E + ++VG+TL +L+ F E L +G+D++ P + G+
Sbjct 534 SMSKKDRKERQMSKKLEEHQRRMTVGTTLGELTAFSESTLDALGDDEEDP----TLGRAN 589
Query 111 KEKTRGRGMLSHYLNAAKQAAEENKKLNKAF-AEKLVIARQQNLQHLN 157
K K + + L+ YLNAAKQA+E ++ A A++ +++RQ+N + L+
Sbjct 590 KSKKKQKNALNAYLNAAKQASETARRAEAALQADRAILSRQENQKTLS 637
> pfa:PF13_0181 conserved Plasmodium protein, unknown function
Length=871
Score = 32.0 bits (71), Expect = 1.0, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 134 NKKLNKAFAEKLVIARQQNLQHLNRKNHKEMDKSAPKKNDSRVGR 178
N K NK K I++Q N+ + N K + + + P N SR+ R
Sbjct 384 NIKKNKEMMNKTNISKQTNISNNNHKKYDTFNNNIPIHNTSRISR 428
> pfa:PFC1015c conserved Plasmodium protein, unknown function
Length=2340
Score = 30.8 bits (68), Expect = 2.5, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 26/49 (53%), Gaps = 10/49 (20%)
Query 127 AKQAAEENKKLNKAFAEKLVIARQQNLQHLNRKNHKEMDKSAPKKNDSR 175
KQ A++N K N +QN + +++NHK+ DK K+ND +
Sbjct 787 GKQNAKQNDKHND----------KQNDKQNDKQNHKQYDKQNDKQNDKQ 825
> sce:YML088W UFO1; F-box receptor protein, subunit of the Skp1-Cdc53-F-box
receptor (SCF) E3 ubiquitin ligase complex; binds
to phosphorylated Ho endonuclease, allowing its ubiquitylation
by SCF and subsequent degradation; K15073 ubiquitin ligase
complex F-box protein UFO1
Length=668
Score = 30.0 bits (66), Expect = 5.0, Method: Composition-based stats.
Identities = 25/53 (47%), Positives = 33/53 (62%), Gaps = 6/53 (11%)
Query 10 EMTGDAPEEVGIERWLAAKAHKEA-AR---EAGL-VGDDEDMQLMRRSLSKKE 57
E++GD EE I+ +A +EA AR EAG VGDDED QL RR+L + +
Sbjct 509 ELSGDTDEENDIQLRIALLESQEAQARNQAEAGEPVGDDEDEQL-RRALEESQ 560
> ath:AT1G53470 MSL4; MSL4 (MECHANOSENSITIVE CHANNEL OF SMALL
CONDUCTANCE-LIKE 4)
Length=881
Score = 29.3 bits (64), Expect = 7.6, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 22/41 (53%), Gaps = 2/41 (4%)
Query 95 GEDDDGPFDLQSRG--KGKEKTRGRGMLSHYLNAAKQAAEE 133
GE+DDG FD R K +E +++ +LN K + +E
Sbjct 47 GEEDDGSFDFMRRSSEKSEEPDPPSKLINQFLNKQKASGDE 87
> ath:AT2G32050 cell cycle control protein-related
Length=254
Score = 28.9 bits (63), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 21/48 (43%), Positives = 29/48 (60%), Gaps = 4/48 (8%)
Query 41 GDDEDMQLMRRSL-SKKERKVQAA---QRALFERRAALSVGSTLEDLS 84
G D L +R+L SK+E V AA +++ RR ++SV S LEDLS
Sbjct 120 GGDRMSSLEKRTLVSKREVDVMAALDEMKSMKSRRVSVSVDSMLEDLS 167
> sce:YOR141C ARP8; Arp8p; K11673 actin-related protein 8
Length=881
Score = 28.9 bits (63), Expect = 9.1, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 26/44 (59%), Gaps = 4/44 (9%)
Query 122 HYLNAAKQAAEENKKLNKA-FAEKLV---IARQQNLQHLNRKNH 161
HY+NA + +ENK++ +A EK+V + QN++H N H
Sbjct 780 HYVNAPDKTEDENKQILQAQIKEKIVEELEEQHQNIEHQNGNEH 823
> tgo:TGME49_060220 folate/methotrexate transporter, putative
Length=1056
Score = 28.9 bits (63), Expect = 9.3, Method: Composition-based stats.
Identities = 25/97 (25%), Positives = 45/97 (46%), Gaps = 11/97 (11%)
Query 28 KAHKEAAREAGLVGDDEDMQLMRRSLSKKER--KVQAAQRALFERRAA--------LSVG 77
+ EA EAG+ D+ + RR++ ++ R KV + AL E R + G
Sbjct 525 RRESEAGAEAGVSSDEATRDVHRRTVERRTRGGKVAETELALMETRGENRTRARENRTFG 584
Query 78 STLED-LSIFCEEELGGVGEDDDGPFDLQSRGKGKEK 113
+ + LS +E+ GG ++D+ + R G+E+
Sbjct 585 ACVHHALSTTDDEDSGGYRDEDEFDATRKPRIFGRER 621
Lambda K H
0.312 0.128 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4795148792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40