bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2414_orf2
Length=267
Score E
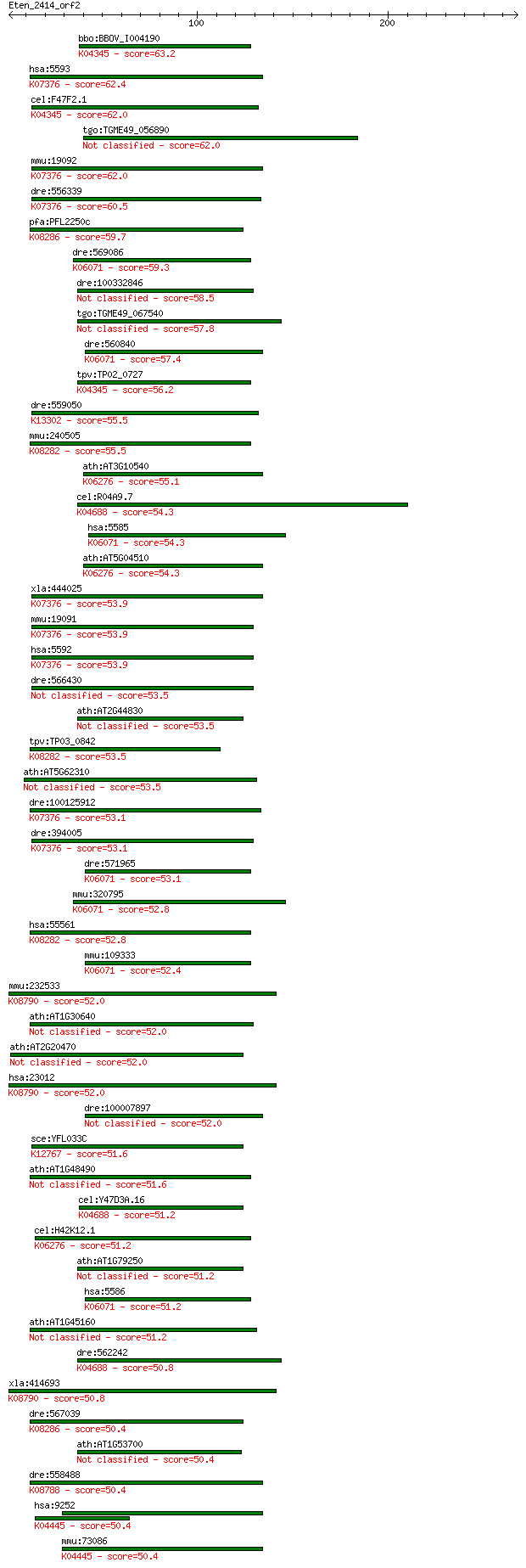
Sequences producing significant alignments: (Bits) Value
bbo:BBOV_I004190 19.m02221; cAMP-dependent protein kinase (EC:... 63.2 1e-09
hsa:5593 PRKG2, PRKGR2, cGKII; protein kinase, cGMP-dependent,... 62.4 2e-09
cel:F47F2.1 hypothetical protein; K04345 protein kinase A [EC:... 62.0 2e-09
tgo:TGME49_056890 protein kinase domain-containing protein (EC... 62.0 2e-09
mmu:19092 Prkg2, AW212535, CGKII, MGC130517, Prkgr2, cGK-II; p... 62.0 2e-09
dre:556339 protein kinase, cGMP-dependent, type II-like; K0737... 60.5 6e-09
pfa:PFL2250c PKB; rac-beta serine/threonine protein kinase, Pf... 59.7 9e-09
dre:569086 pkn2; protein kinase N2; K06071 protein kinase N [E... 59.3 1e-08
dre:100332846 acyl-CoA synthetase long-chain family member 6-like 58.5 3e-08
tgo:TGME49_067540 protein kinase, putative (EC:4.1.1.70 2.7.11... 57.8 5e-08
dre:560840 hypothetical LOC560840; K06071 protein kinase N [EC... 57.4 6e-08
tpv:TP02_0727 cAMP-dependent protein kinase catalytic subunit;... 56.2 1e-07
dre:559050 sgk2b, MGC154065, zgc:154065; serum/glucocorticoid ... 55.5 2e-07
mmu:240505 Cdc42bpg, BC046418, MRCKgamma; CDC42 binding protei... 55.5 2e-07
ath:AT3G10540 3-phosphoinositide-dependent protein kinase, put... 55.1 3e-07
cel:R04A9.7 hypothetical protein; K04688 p70 ribosomal S6 kina... 54.3 4e-07
hsa:5585 PKN1, DBK, MGC46204, PAK-1, PAK1, PKN, PKN-ALPHA, PRK... 54.3 5e-07
ath:AT5G04510 PDK1; PDK1 (3'-PHOSPHOINOSITIDE-DEPENDENT PROTEI... 54.3 5e-07
xla:444025 prkg2, MGC82580; protein kinase, cGMP-dependent, ty... 53.9 5e-07
mmu:19091 Prkg1, AW125416, CGKI, MGC132849, Prkg1b, Prkgr1b; p... 53.9 6e-07
hsa:5592 PRKG1, CGKI, DKFZp686K042, FLJ36117, MGC71944, PGK, P... 53.9 6e-07
dre:566430 cGMP dependent protein kinase I 53.5 7e-07
ath:AT2G44830 protein kinase, putative 53.5 7e-07
tpv:TP03_0842 protein kinase (EC:2.7.1.37); K08282 non-specifi... 53.5 7e-07
ath:AT5G62310 IRE; IRE (INCOMPLETE ROOT HAIR ELONGATION); kina... 53.5 8e-07
dre:100125912 prkg2; protein kinase, cGMP-dependent, type II (... 53.1 1e-06
dre:394005 prkg1a, MGC63955, prkg1, zgc:63955; protein kinase,... 53.1 1e-06
dre:571965 protein kinase N2-like; K06071 protein kinase N [EC... 53.1 1e-06
mmu:320795 Pkn1, DBK, F730027O18Rik, PAK1, PRK1, Pkn, Prkcl1, ... 52.8 1e-06
hsa:55561 CDC42BPG, DMPK2, HSMDPKIN, KAPPA-200, MRCKgamma; CDC... 52.8 1e-06
mmu:109333 Pkn2, 6030436C20Rik, AI507382, PRK2, Prkcl2, Stk7; ... 52.4 2e-06
mmu:232533 Stk38l, 4930473A22Rik, B230328I19, Ndr2, Ndr54; ser... 52.0 2e-06
ath:AT1G30640 protein kinase, putative 52.0 2e-06
ath:AT2G20470 kinase 52.0 2e-06
hsa:23012 STK38L, KIAA0965, NDR2; serine/threonine kinase 38 l... 52.0 2e-06
dre:100007897 MGC162290; zgc:162290 52.0 3e-06
sce:YFL033C RIM15, TAK1; Glucose-repressible protein kinase in... 51.6 3e-06
ath:AT1G48490 kinase 51.6 3e-06
cel:Y47D3A.16 rsks-1; RSK-pSeventy (RSK-p70 kinase) homolog fa... 51.2 3e-06
cel:H42K12.1 pdk-1; PDK-class protein kinase family member (pd... 51.2 3e-06
ath:AT1G79250 AGC1.7; AGC1.7 (AGC KINASE 1.7); kinase 51.2 4e-06
hsa:5586 PKN2, MGC150606, MGC71074, PAK2, PRK2, PRKCL2, PRO204... 51.2 4e-06
ath:AT1G45160 kinase 51.2 4e-06
dre:562242 ribosomal protein S6 kinase b, polypeptide 1-like; ... 50.8 4e-06
xla:414693 stk38l, MGC83214, ndr2; serine/threonine kinase 38 ... 50.8 5e-06
dre:567039 cdc42bpb, si:dkeyp-93h6.2, wu:fb98c02, wu:fc48b05, ... 50.4 6e-06
ath:AT1G53700 WAG1; WAG1 (WAG 1); kinase/ protein serine/threo... 50.4 6e-06
dre:558488 serine/threonine-protein kinase MRCK alpha-like; K0... 50.4 6e-06
hsa:9252 RPS6KA5, MGC1911, MSK1, MSPK1, RLPK; ribosomal protei... 50.4 6e-06
mmu:73086 Rps6ka5, 3110005L17Rik, 6330404E13Rik, AI854034, MGC... 50.4 7e-06
> bbo:BBOV_I004190 19.m02221; cAMP-dependent protein kinase (EC:2.7.11.1);
K04345 protein kinase A [EC:2.7.11.11]
Length=416
Score = 63.2 bits (152), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 36/93 (38%), Positives = 49/93 (52%), Gaps = 5/93 (5%)
Query 38 CGYGT--DCWSLGCLVHELLVGEPPFKSVSSTAVAQ-ALCGYHELSLTQQMPAAAVDFIK 94
CG+G D WSLG LV+E+L G PPF + +A AL + L+ ++ DFI+
Sbjct 278 CGHGIEADWWSLGVLVYEILTGVPPFHASDPSATYDLALANHIRFPLSVRVEVK--DFIR 335
Query 95 SLLRPKEEERLGSRDIKELLKHHFLEGVDCMAL 127
LL +RLG + EL H F G+D M L
Sbjct 336 RLLVVNPRKRLGGSNSSELYSHRFFAGMDWMKL 368
> hsa:5593 PRKG2, PRKGR2, cGKII; protein kinase, cGMP-dependent,
type II (EC:2.7.11.12); K07376 protein kinase, cGMP-dependent
[EC:2.7.11.12]
Length=762
Score = 62.4 bits (150), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 39/125 (31%), Positives = 55/125 (44%), Gaps = 15/125 (12%)
Query 12 VPGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ 71
P Y APE ++ D + D WSLG LV+ELL G PPF V
Sbjct 613 TPEYVAPEVILNKGHD------------FSVDFWSLGILVYELLTGNPPFSGVDQMMTYN 660
Query 72 -ALCGYHELSLTQQMPAAAVDFIKSLLRPKEEERLGS--RDIKELLKHHFLEGVDCMALH 128
L G ++ +++ D I+ L R ERLG+ I ++ KH +L G + L
Sbjct 661 LILKGIEKMDFPRKITRRPEDLIRRLCRQNPTERLGNLKNGINDIKKHRWLNGFNWEGLK 720
Query 129 FQPLP 133
+ LP
Sbjct 721 ARSLP 725
> cel:F47F2.1 hypothetical protein; K04345 protein kinase A [EC:2.7.11.11]
Length=398
Score = 62.0 bits (149), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/127 (29%), Positives = 62/127 (48%), Gaps = 24/127 (18%)
Query 13 PGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQA 72
P Y APE+ G G D W+LG L++E++VG+PPF+ +++ + A
Sbjct 249 PDYLAPESLART------------GHNKGVDWWALGILIYEMMVGKPPFRGKTTSEIYDA 296
Query 73 LCGYHELSLTQQMPAAAVDFIKSLLRPKEEERL-----GSRDIKELLKHHFLEGV---DC 124
+ H+L + AA D +K LL +R+ G++D+K+ H + E V D
Sbjct 297 IIE-HKLKFPRSFNLAAKDLVKKLLEVDRTQRIGCMKNGTQDVKD---HKWFEKVNWDDT 352
Query 125 MALHFQP 131
+ L +P
Sbjct 353 LHLRVEP 359
> tgo:TGME49_056890 protein kinase domain-containing protein (EC:2.7.11.1
2.7.11.11)
Length=1590
Score = 62.0 bits (149), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 49/146 (33%), Positives = 67/146 (45%), Gaps = 4/146 (2%)
Query 40 YGTDCWSLGCLVHELLVGEPPFKS-VSSTAVAQALCGYHELSLTQQMPAAAVDFIKSLLR 98
Y D WSLGC+ +E+L G P F++ V V + L G E + A A DFI LL
Sbjct 812 YALDLWSLGCIAYEMLCGRPAFEAQVPQETVEKILRGAVE--FPSSLSAEARDFISRLLV 869
Query 99 PKEEERLGSRDIKELLKHHFLEGVDCMALHFQPLPLDVMQYDFAFQGFGHSCRLGATAAK 158
+ ERLG RD EL H FL + +PL + + + + + AK
Sbjct 870 VEPTERLGFRDFSELKFHPFLASLGEDFWRLPSVPLVRLYERKRVRTLRGAEKANSREAK 929
Query 159 CDCTVPVEREHNAVR-AAASAAGTPS 183
+ RE +A+R A AS AG S
Sbjct 930 REPESESAREADALREATASNAGQSS 955
> mmu:19092 Prkg2, AW212535, CGKII, MGC130517, Prkgr2, cGK-II;
protein kinase, cGMP-dependent, type II (EC:2.7.11.12); K07376
protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=762
Score = 62.0 bits (149), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 38/124 (30%), Positives = 55/124 (44%), Gaps = 15/124 (12%)
Query 13 PGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ- 71
P Y APE ++ D + D WSLG LV+ELL G PPF +
Sbjct 614 PEYVAPEVILNKGHD------------FSVDFWSLGILVYELLTGNPPFSGIDQMMTYNL 661
Query 72 ALCGYHELSLTQQMPAAAVDFIKSLLRPKEEERLGS--RDIKELLKHHFLEGVDCMALHF 129
L G ++ +++ D I+ L R ERLG+ I ++ KH +L G + L
Sbjct 662 ILKGIEKMDFPRKITRRPEDLIRRLCRQNPTERLGNLKNGINDIKKHRWLNGFNWEGLKA 721
Query 130 QPLP 133
+ LP
Sbjct 722 RSLP 725
> dre:556339 protein kinase, cGMP-dependent, type II-like; K07376
protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=730
Score = 60.5 bits (145), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 36/123 (29%), Positives = 58/123 (47%), Gaps = 15/123 (12%)
Query 13 PGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAV-AQ 71
P Y APE ++ D +G DCWSLG L+ ELL+G PPF +
Sbjct 582 PEYVAPEVIMNKGHD------------FGADCWSLGILIFELLIGSPPFTGSDPIRIYTM 629
Query 72 ALCGYHELSLTQQMPAAAVDFIKSLLRPKEEERLGSRD--IKELLKHHFLEGVDCMALHF 129
L G ++ + +++ D I+ L + ERLG++ I ++ KH + +G + L
Sbjct 630 VLHGIEKVDIPKRISKRPEDLIRRLCKLNPAERLGNKKNGIIDIKKHKWFQGFNWEGLRR 689
Query 130 QPL 132
+ L
Sbjct 690 RKL 692
> pfa:PFL2250c PKB; rac-beta serine/threonine protein kinase,
PfPKB (EC:2.7.1.-); K08286 protein-serine/threonine kinase [EC:2.7.11.-]
Length=735
Score = 59.7 bits (143), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 34/114 (29%), Positives = 57/114 (50%), Gaps = 15/114 (13%)
Query 12 VPGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ 71
P Y APE + Q G G D WSLG +++E+L GE PF + + + +
Sbjct 564 TPEYLAPEI----------IEQK--GHGKAVDWWSLGIMLYEMLTGELPFNNTNRNVLFE 611
Query 72 ALCGYHELSLTQQMPAAAVDFIKSLLRPKEEERLGS--RDIKELLKHHFLEGVD 123
++ Y +L+ + + AVD + L ++RLGS D +E+ KH F + ++
Sbjct 612 SI-KYQKLNYPKNLSPKAVDLLTKLFEKNPKKRLGSGGTDAQEIKKHPFFKNIN 664
> dre:569086 pkn2; protein kinase N2; K06071 protein kinase N
[EC:2.7.11.13]
Length=940
Score = 59.3 bits (142), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 49/95 (51%), Gaps = 3/95 (3%)
Query 35 DFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIK 94
D D W LG L++E+LVGE PF V ++ E+ + M +V I+
Sbjct 788 DSNYTRAVDWWGLGVLIYEMLVGESPFPGDDEEEVFDSIVN-DEVRYPRFMSPESVSVIQ 846
Query 95 SLLRPKEEERLGS--RDIKELLKHHFLEGVDCMAL 127
LL+ E+RLG+ +D E+ KH F +G+D AL
Sbjct 847 KLLQKNSEKRLGAGEQDANEVKKHRFFQGIDWEAL 881
> dre:100332846 acyl-CoA synthetase long-chain family member 6-like
Length=1391
Score = 58.5 bits (140), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 47/94 (50%), Gaps = 2/94 (2%)
Query 37 GCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQA-LCGYHELSLTQQMPAAAVDFIKS 95
G D W+LG L++ELL G P F V + L G EL + + A IKS
Sbjct 566 GHSLSADLWALGMLLYELLNGSPLFSDSEHMKVYRGTLKGTDELEFPKTISRTAAHLIKS 625
Query 96 LLRPKEEERLGSRD-IKELLKHHFLEGVDCMALH 128
L R K ERLG R+ +K++ +H + EG D L
Sbjct 626 LCRLKPTERLGQRNGVKDVCRHMWFEGFDWEGLQ 659
> tgo:TGME49_067540 protein kinase, putative (EC:4.1.1.70 2.7.11.13)
Length=951
Score = 57.8 bits (138), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 38/119 (31%), Positives = 54/119 (45%), Gaps = 13/119 (10%)
Query 37 GCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIKSL 96
G G D WSLG L++E+L G PPF + + + + EL M AAV ++ L
Sbjct 633 GHGKAVDWWSLGALIYEMLTGLPPFYTGDRERLFENIRS-SELQYPSYMSRAAVHLLRGL 691
Query 97 LRPKEEERLGS--RDIKELLKHHFLEGVDCMALH-------FQPL---PLDVMQYDFAF 143
+ RLG D +E+ +H F +D AL F+P P DV +D F
Sbjct 692 FQRDPNRRLGGGPGDAEEIKRHPFFGRIDWEALQAKRMRPPFRPRLQSPTDVQYFDNEF 750
> dre:560840 hypothetical LOC560840; K06071 protein kinase N [EC:2.7.11.13]
Length=948
Score = 57.4 bits (137), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 49/95 (51%), Gaps = 3/95 (3%)
Query 41 GTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIKSLLRPK 100
D W LG L++E+LVGE PF V ++ E+ + + A A+ ++ LLR
Sbjct 801 AVDWWGLGVLIYEMLVGESPFPGDDEEEVFDSIVN-DEVRYPRFLSAEAIGIMRRLLRRN 859
Query 101 EEERLGS--RDIKELLKHHFLEGVDCMALHFQPLP 133
E RLGS +D +++ K F +D AL + LP
Sbjct 860 PERRLGSSEKDAEDVKKQPFFRNMDWEALLLRKLP 894
> tpv:TP02_0727 cAMP-dependent protein kinase catalytic subunit;
K04345 protein kinase A [EC:2.7.11.11]
Length=511
Score = 56.2 bits (134), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 31/91 (34%), Positives = 51/91 (56%), Gaps = 2/91 (2%)
Query 37 GCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIKSL 96
G + D +S G ++ELL G PPF S +S L +E++ +++ + D IK+L
Sbjct 376 GHDFRADFYSFGVFLYELLTGVPPFYS-NSPQKTYKLALNNEVAFGRKVNNVSRDIIKNL 434
Query 97 LRPKEEERLGSRDIKELLKHHFLEGVDCMAL 127
LR +RLG+ ++KE+ H F +G+D L
Sbjct 435 LRVDPSKRLGN-NVKEVYFHPFFDGIDFNML 464
> dre:559050 sgk2b, MGC154065, zgc:154065; serum/glucocorticoid
regulated kinase 2b; K13302 serum/glucocorticoid-regulated
kinase 1 [EC:2.7.11.1]
Length=404
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 38/123 (30%), Positives = 57/123 (46%), Gaps = 17/123 (13%)
Query 13 PGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQA 72
P Y APE + + D D W LG ++HE+L G PPF + + +
Sbjct 240 PEYLAPEVLLQEEYDRT------------VDWWGLGAVLHEMLYGLPPFYNADRLEMLRN 287
Query 73 LCGYHELSLTQQMPAAAVDFIKSLLRPKEEERLGS-RDIKELLKHHFLEGV---DCMALH 128
+ Y L+L + +AA D +K LL +RLG+ RD+ EL H F + + +A
Sbjct 288 II-YQPLALKAGVSSAARDLLKRLLNRDRAKRLGAKRDLIELQSHAFFSPIQWDELVAKK 346
Query 129 FQP 131
QP
Sbjct 347 IQP 349
> mmu:240505 Cdc42bpg, BC046418, MRCKgamma; CDC42 binding protein
kinase gamma (DMPK-like) (EC:2.7.11.1); K08282 non-specific
serine/threonine protein kinase [EC:2.7.11.1]
Length=1551
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 42/121 (34%), Positives = 57/121 (47%), Gaps = 14/121 (11%)
Query 12 VPGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ 71
P Y +PE +++ + Y C D WSLG +ELL GE PF + S
Sbjct 234 TPDYISPEILQAME---EGKGHYGPQC----DWWSLGVCAYELLFGETPFYAESLVETYG 286
Query 72 ALCGYHELSL-----TQQMPAAAVDFIKSLLRPKEEERLGSRDIKELLKHHFLEGVDCMA 126
+ HE L +PA+A D I+ LL ++EERLG + + KH F EGVD
Sbjct 287 KIMN-HEDHLQFPADVTDVPASAQDLIRQLL-CRQEERLGRGGLDDFRKHPFFEGVDWER 344
Query 127 L 127
L
Sbjct 345 L 345
> ath:AT3G10540 3-phosphoinositide-dependent protein kinase, putative;
K06276 3-phosphoinositide dependent protein kinase-1
[EC:2.7.11.1]
Length=486
Score = 55.1 bits (131), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 43/96 (44%), Gaps = 3/96 (3%)
Query 40 YGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIKSLLRP 99
+G D W+LGC ++++L G PFK S + Q + ++ AA D I LL
Sbjct 232 FGNDLWALGCTLYQMLSGTSPFKDASEWLIFQRIIA-RDIKFPNHFSEAARDLIDRLLDT 290
Query 100 KEEER--LGSRDIKELLKHHFLEGVDCMALHFQPLP 133
R GS L +H F +GVD L Q P
Sbjct 291 DPSRRPGAGSEGYDSLKRHPFFKGVDWKNLRSQTPP 326
> cel:R04A9.7 hypothetical protein; K04688 p70 ribosomal S6 kinase
[EC:2.7.11.1]
Length=402
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 45/184 (24%), Positives = 81/184 (44%), Gaps = 23/184 (12%)
Query 37 GCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIKSL 96
G + D W+LG L++++ +G PPF + +++ ++ L ++ A+ D IK +
Sbjct 227 GHDHAVDVWALGILMYDMFMGGPPFTGETPADKDKSILK-GKVRLPPKLSASGKDLIKRI 285
Query 97 LRPKEEERLGSRDIKELLKHHFLEGVD---CMALHFQPLPL--------DVMQYDFAFQG 145
++ R+ DIKE H F E +D MA F+P P DV +D +F
Sbjct 286 IKRDPTLRITIPDIKE---HEFFEEIDWDKLMAHDFEP-PFKPTLANLEDVSHFDESFTS 341
Query 146 FGHSCRLGATAAKCDCTVPVEREHNAVRAAASAAGTPSALETPVTSLSGSCVSTSKSIAS 205
+ + C V E N + + T ET T+ + ++T+ ++ +
Sbjct 342 ------IAPEESPCKAIVTENEERNYLFNGFNFNRTSKTKETRKTTSAARKLTTTNNVVT 395
Query 206 DAPQ 209
APQ
Sbjct 396 -APQ 398
> hsa:5585 PKN1, DBK, MGC46204, PAK-1, PAK1, PKN, PKN-ALPHA, PRK1,
PRKCL1; protein kinase N1 (EC:2.7.11.13); K06071 protein
kinase N [EC:2.7.11.13]
Length=942
Score = 54.3 bits (129), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 54/115 (46%), Gaps = 13/115 (11%)
Query 43 DCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIKSLLRPKEE 102
D W LG L++E+LVGE PF V ++ E+ + + A A+ ++ LLR E
Sbjct 797 DWWGLGVLLYEMLVGESPFPGDDEEEVFDSIVN-DEVRYPRFLSAEAIGIMRRLLRRNPE 855
Query 103 ERLGS--RDIKELLKHHFLEGVDCMALHFQPLP----------LDVMQYDFAFQG 145
RLGS RD +++ K F + AL + LP DV +D F G
Sbjct 856 RRLGSSERDAEDVKKQPFFRTLGWEALLARRLPPPFVPTLSGRTDVSNFDEEFTG 910
> ath:AT5G04510 PDK1; PDK1 (3'-PHOSPHOINOSITIDE-DEPENDENT PROTEIN
KINASE 1); 3-phosphoinositide-dependent protein kinase/
kinase/ phosphoinositide binding / protein binding / protein
kinase; K06276 3-phosphoinositide dependent protein kinase-1
[EC:2.7.11.1]
Length=491
Score = 54.3 bits (129), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 43/96 (44%), Gaps = 3/96 (3%)
Query 40 YGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIKSLLRP 99
+G D W+LGC ++++L G PFK S + Q + ++ AA D I LL
Sbjct 231 FGNDLWALGCTLYQMLSGTSPFKDASEWLIFQRIIA-RDIKFPNHFSEAARDLIDRLLDT 289
Query 100 KEEER--LGSRDIKELLKHHFLEGVDCMALHFQPLP 133
+ R GS L +H F GVD L Q P
Sbjct 290 EPSRRPGAGSEGYVALKRHPFFNGVDWKNLRSQTPP 325
> xla:444025 prkg2, MGC82580; protein kinase, cGMP-dependent,
type II (EC:2.7.11.1); K07376 protein kinase, cGMP-dependent
[EC:2.7.11.12]
Length=783
Score = 53.9 bits (128), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 52/124 (41%), Gaps = 15/124 (12%)
Query 13 PGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ- 71
P Y APE ++ G + D WSLG L++ELL G PPF +
Sbjct 635 PEYVAPEVILNK------------GHSFSVDFWSLGILLYELLTGNPPFTGPDQMIIYNL 682
Query 72 ALCGYHELSLTQQMPAAAVDFIKSLLRPKEEERLG--SRDIKELLKHHFLEGVDCMALHF 129
L G ++ + + D I L R ERLG I ++ KH + G + L+
Sbjct 683 ILQGIEKIEFCKNITKRPEDLICRLCRQNPAERLGYMKNGIADIKKHRWFNGFNWEGLNT 742
Query 130 QPLP 133
+ LP
Sbjct 743 RSLP 746
> mmu:19091 Prkg1, AW125416, CGKI, MGC132849, Prkg1b, Prkgr1b;
protein kinase, cGMP-dependent, type I (EC:2.7.11.12); K07376
protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=671
Score = 53.9 bits (128), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 51/119 (42%), Gaps = 15/119 (12%)
Query 13 PGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ- 71
P Y APE ++ G D WSLG L++ELL G PPF
Sbjct 522 PEYVAPEIILNK------------GHDISADYWSLGILMYELLTGSPPFSGPDPMKTYNI 569
Query 72 ALCGYHELSLTQQMPAAAVDFIKSLLRPKEEERLGS--RDIKELLKHHFLEGVDCMALH 128
L G + +++ A + IK L R ERLG+ +K++ KH + EG + L
Sbjct 570 ILRGIDMIEFPKKIAKNAANLIKKLCRDNPSERLGNLKNGVKDIQKHKWFEGFNWEGLR 628
> hsa:5592 PRKG1, CGKI, DKFZp686K042, FLJ36117, MGC71944, PGK,
PKG, PRKG1B, PRKGR1B, cGKI-BETA, cGKI-alpha; protein kinase,
cGMP-dependent, type I (EC:2.7.11.12); K07376 protein kinase,
cGMP-dependent [EC:2.7.11.12]
Length=671
Score = 53.9 bits (128), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 51/119 (42%), Gaps = 15/119 (12%)
Query 13 PGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ- 71
P Y APE ++ D D WSLG L++ELL G PPF
Sbjct 522 PEYVAPEIILNKGHD------------ISADYWSLGILMYELLTGSPPFSGPDPMKTYNI 569
Query 72 ALCGYHELSLTQQMPAAAVDFIKSLLRPKEEERLGS--RDIKELLKHHFLEGVDCMALH 128
L G + +++ A + IK L R ERLG+ +K++ KH + EG + L
Sbjct 570 ILRGIDMIEFPKKIAKNAANLIKKLCRDNPSERLGNLKNGVKDIQKHKWFEGFNWEGLR 628
> dre:566430 cGMP dependent protein kinase I
Length=528
Score = 53.5 bits (127), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 51/119 (42%), Gaps = 15/119 (12%)
Query 13 PGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ- 71
P Y APE ++ D D WSLG L++ELL G PPF
Sbjct 379 PEYVAPEIILNKGHD------------ISADYWSLGILMYELLTGSPPFSGPDPMKTYNI 426
Query 72 ALCGYHELSLTQQMPAAAVDFIKSLLRPKEEERLGS--RDIKELLKHHFLEGVDCMALH 128
L G + +++ A + IK L R ERLG+ +K++ KH + EG + L
Sbjct 427 ILRGIDMIEFPKKITKNAGNLIKKLCRDNPSERLGNLKNGVKDIQKHKWFEGFNWEGLR 485
> ath:AT2G44830 protein kinase, putative
Length=765
Score = 53.5 bits (127), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 34/91 (37%), Positives = 47/91 (51%), Gaps = 6/91 (6%)
Query 37 GCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPA---AAVDFI 93
G G D W+ G VHELL G+ PFK + A + G +L + PA A D I
Sbjct 613 GHGSAVDWWTFGIFVHELLYGKTPFKGSGNRATLFNVVG-EQLKFPES-PATSYAGRDLI 670
Query 94 KSLLRPKEEERLGS-RDIKELLKHHFLEGVD 123
++LL + RLG+ R E+ +H F EGV+
Sbjct 671 QALLVKDPKNRLGTKRGATEIKQHPFFEGVN 701
> tpv:TP03_0842 protein kinase (EC:2.7.1.37); K08282 non-specific
serine/threonine protein kinase [EC:2.7.11.1]
Length=417
Score = 53.5 bits (127), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 34/100 (34%), Positives = 49/100 (49%), Gaps = 14/100 (14%)
Query 12 VPGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ 71
P + PEA + DS G+ D WSLGCL++++LVG PPF + +
Sbjct 229 TPNFMDPEAISNADS------------GFKRDFWSLGCLMYQVLVGRPPFYGSTEYFIID 276
Query 72 ALCGYHELSLTQQMPAAAVDFIKSLLRPKEEERLGSRDIK 111
+ + +LS + A D I SLLR E+RLG I+
Sbjct 277 RVRKF-DLSFPPSINPDAKDLILSLLRDP-EDRLGFDQIR 314
> ath:AT5G62310 IRE; IRE (INCOMPLETE ROOT HAIR ELONGATION); kinase/
protein serine/threonine kinase
Length=1168
Score = 53.5 bits (127), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 36/126 (28%), Positives = 59/126 (46%), Gaps = 16/126 (12%)
Query 9 HAVV--PGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSS 66
HAVV P Y APE + + G G D WS+G ++ E+LVG PPF + +
Sbjct 941 HAVVGTPDYLAPEILLGM------------GHGKTADWWSVGVILFEVLVGIPPFNAETP 988
Query 67 TAVAQALCGYH--ELSLTQQMPAAAVDFIKSLLRPKEEERLGSRDIKELLKHHFLEGVDC 124
+ + + ++ +++ A D I LL +RLG+ E+ +HHF + ++
Sbjct 989 QQIFENIINRDIPWPNVPEEISYEAHDLINKLLTENPVQRLGATGAGEVKQHHFFKDINW 1048
Query 125 MALHFQ 130
L Q
Sbjct 1049 DTLARQ 1054
> dre:100125912 prkg2; protein kinase, cGMP-dependent, type II
(EC:2.7.11.1); K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=768
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 51/124 (41%), Gaps = 15/124 (12%)
Query 12 VPGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ 71
P Y APE ++ G G D WSLG L+ ELL G PPF +
Sbjct 619 TPEYVAPEIILNK------------GHGLSVDFWSLGILIFELLTGSPPFTGSDQMIIYT 666
Query 72 -ALCGYHELSLTQQMPAAAVDFIKSLLRPKEEERLGS--RDIKELLKHHFLEGVDCMALH 128
L G ++ +++ D I+ L R ERLG+ I ++ KH + G L
Sbjct 667 FILKGIEKMDFPKKITKRPGDLIRKLCRQNPSERLGNLKNGITDIKKHRWFTGFSWSGLK 726
Query 129 FQPL 132
+ L
Sbjct 727 ARNL 730
> dre:394005 prkg1a, MGC63955, prkg1, zgc:63955; protein kinase,
cGMP-dependent, type Ia (EC:2.7.11.1); K07376 protein kinase,
cGMP-dependent [EC:2.7.11.12]
Length=667
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 51/119 (42%), Gaps = 15/119 (12%)
Query 13 PGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ- 71
P Y APE ++ G D WSLG L++ELL G PPF
Sbjct 518 PEYVAPEIILNK------------GHDISADYWSLGILMYELLTGSPPFSGPDPMKTYNI 565
Query 72 ALCGYHELSLTQQMPAAAVDFIKSLLRPKEEERLGS--RDIKELLKHHFLEGVDCMALH 128
L G + +++ A + IK L R ERLG+ +K++ KH + EG + L
Sbjct 566 ILRGIDMIEFPKKITKNAANLIKKLCRDTPSERLGNLKNGVKDIQKHKWFEGFNWDGLR 624
> dre:571965 protein kinase N2-like; K06071 protein kinase N [EC:2.7.11.13]
Length=970
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 44/89 (49%), Gaps = 3/89 (3%)
Query 41 GTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIKSLLRPK 100
D W LG L+ E+LVGE PF V ++ E+ + + A+ ++ LLR
Sbjct 823 AVDWWGLGVLIFEMLVGESPFPGDDEEEVFDSIVN-DEVRYPRVLSTEAISIMRRLLRRN 881
Query 101 EEERLGS--RDIKELLKHHFLEGVDCMAL 127
E RLG+ RD +++ KH F +D L
Sbjct 882 PERRLGAAERDAEDVKKHLFFRDIDWDGL 910
> mmu:320795 Pkn1, DBK, F730027O18Rik, PAK1, PRK1, Pkn, Prkcl1,
Stk3; protein kinase N1 (EC:2.7.11.13); K06071 protein kinase
N [EC:2.7.11.13]
Length=951
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 38/123 (30%), Positives = 54/123 (43%), Gaps = 13/123 (10%)
Query 35 DFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIK 94
D D W LG L++E+LVGE PF V ++ E+ + + A A+ ++
Sbjct 798 DTSYTRAVDWWGLGVLLYEMLVGESPFPGDDEEEVFDSIVN-DEVRYPRFLSAEAIGIMR 856
Query 95 SLLRPKEEERLGS--RDIKELLKHHFLE--GVDCMALHFQPLPL--------DVMQYDFA 142
LLR E RLGS RD +++ K F G D + P P DV +D
Sbjct 857 RLLRRNPERRLGSTERDAEDVKKQPFFRSLGWDVLLARRLPPPFVPTLSGRTDVSNFDEE 916
Query 143 FQG 145
F G
Sbjct 917 FTG 919
> hsa:55561 CDC42BPG, DMPK2, HSMDPKIN, KAPPA-200, MRCKgamma; CDC42
binding protein kinase gamma (DMPK-like) (EC:2.7.11.1);
K08282 non-specific serine/threonine protein kinase [EC:2.7.11.1]
Length=1551
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 41/121 (33%), Positives = 56/121 (46%), Gaps = 14/121 (11%)
Query 12 VPGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ 71
P Y +PE +++ + Y C D WSLG +ELL GE PF + S
Sbjct 234 TPDYISPEILQAME---EGKGHYGPQC----DWWSLGVCAYELLFGETPFYAESLVETYG 286
Query 72 ALCGYHELSL-----TQQMPAAAVDFIKSLLRPKEEERLGSRDIKELLKHHFLEGVDCMA 126
+ HE L +PA+A D I+ LL ++EERLG + + H F EGVD
Sbjct 287 KIMN-HEDHLQFPPDVPDVPASAQDLIRQLL-CRQEERLGRGGLDDFRNHPFFEGVDWER 344
Query 127 L 127
L
Sbjct 345 L 345
> mmu:109333 Pkn2, 6030436C20Rik, AI507382, PRK2, Prkcl2, Stk7;
protein kinase N2 (EC:2.7.11.13); K06071 protein kinase N
[EC:2.7.11.13]
Length=983
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 45/89 (50%), Gaps = 3/89 (3%)
Query 41 GTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIKSLLRPK 100
D W LG L++E+LVGE PF V ++ E+ + + A+ ++ LLR
Sbjct 836 AVDWWGLGVLIYEMLVGESPFPGDDEEEVFDSIVN-DEVRYPRFLSTEAISIMRRLLRRN 894
Query 101 EEERLGS--RDIKELLKHHFLEGVDCMAL 127
E RLG+ +D +++ KH F D AL
Sbjct 895 PERRLGAGEKDAEDVKKHPFFRLTDWSAL 923
> mmu:232533 Stk38l, 4930473A22Rik, B230328I19, Ndr2, Ndr54; serine/threonine
kinase 38 like (EC:2.7.11.1); K08790 serine/threonine
kinase 38 [EC:2.7.11.1]
Length=464
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 44/149 (29%), Positives = 69/149 (46%), Gaps = 28/149 (18%)
Query 1 RQTAVIYPHAVVPGYAAPEASVSVDSDGDSVAQYDFGCGYGT--DCWSLGCLVHELLVGE 58
RQ A Y P Y APE + GY D WSLG +++E+L+G
Sbjct 277 RQLA--YSTVGTPDYIAPEVFMQ--------------TGYNKLCDWWSLGVIMYEMLIGF 320
Query 59 PPFKSVSSTAVAQALCGYHE-LSLTQQMPAA--AVDFIKSLLR--PKEEERLGSRDIKEL 113
PPF S + + + + E L+ ++P + A D I LR E R+G+ ++E+
Sbjct 321 PPFCSETPQETYRKVMSWKETLAFPPEVPVSEKAKDLI---LRFCTDSENRIGNGGVEEI 377
Query 114 LKHHFLEGVDCMALHFQP--LPLDVMQYD 140
H F EGVD + +P +P+++ D
Sbjct 378 KGHPFFEGVDWGHIRERPAAIPIEIRSID 406
> ath:AT1G30640 protein kinase, putative
Length=562
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 55/122 (45%), Gaps = 20/122 (16%)
Query 12 VPGYAAPEASVSVDSDGDSVAQYDFGCGYGTDC--WSLGCLVHELLVGEPPFKSVSSTAV 69
P Y APE + GYG +C WSLG ++ E+LVG PPF S A
Sbjct 326 TPDYIAPEVLLK--------------KGYGMECDWWSLGAIMFEMLVGFPPFYSEEPLAT 371
Query 70 AQALCGYH---ELSLTQQMPAAAVDFIKSLLRPKEEERLGSRDIKELLKHHFLEGVDCMA 126
+ + + + ++ D I+ LL E+RLG++ + E+ H + GV+
Sbjct 372 CRKIVNWKTCLKFPDEAKLSIEVKDLIRRLL-CNVEQRLGTKGVHEIKAHPWFRGVEWER 430
Query 127 LH 128
L+
Sbjct 431 LY 432
> ath:AT2G20470 kinase
Length=569
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 39/127 (30%), Positives = 56/127 (44%), Gaps = 20/127 (15%)
Query 2 QTAVIYPHAVVPGYAAPEASVSVDSDGDSVAQYDFGCGYGTDC--WSLGCLVHELLVGEP 59
+ + Y P Y APE + GYG +C WSLG +++E+LVG P
Sbjct 314 RRTLAYSTVGTPDYIAPEVLLK--------------KGYGMECDWWSLGAIMYEMLVGYP 359
Query 60 PFKSVSSTAVAQALCGYH-ELSLTQQ--MPAAAVDFIKSLLRPKEEERLGSRDIKELLKH 116
PF S + + + + L ++ + A D I SLL RLGS+ EL H
Sbjct 360 PFYSDDPMSTCRKIVNWKSHLKFPEEAILSREAKDLINSLL-CSVRRRLGSKGADELKAH 418
Query 117 HFLEGVD 123
+ E VD
Sbjct 419 TWFETVD 425
> hsa:23012 STK38L, KIAA0965, NDR2; serine/threonine kinase 38
like (EC:2.7.11.1); K08790 serine/threonine kinase 38 [EC:2.7.11.1]
Length=464
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 44/149 (29%), Positives = 68/149 (45%), Gaps = 28/149 (18%)
Query 1 RQTAVIYPHAVVPGYAAPEASVSVDSDGDSVAQYDFGCGYGT--DCWSLGCLVHELLVGE 58
RQ A Y P Y APE + GY D WSLG +++E+L+G
Sbjct 277 RQLA--YSTVGTPDYIAPEVFMQ--------------TGYNKLCDWWSLGVIMYEMLIGY 320
Query 59 PPFKSVSSTAVAQALCGYHE-LSLTQQMPAA--AVDFIKSLLR--PKEEERLGSRDIKEL 113
PPF S + + + + E L ++P + A D I LR E R+G+ ++E+
Sbjct 321 PPFCSETPQETYRKVMNWKETLVFPPEVPISEKAKDLI---LRFCIDSENRIGNSGVEEI 377
Query 114 LKHHFLEGVDCMALHFQP--LPLDVMQYD 140
H F EGVD + +P +P+++ D
Sbjct 378 KGHPFFEGVDWEHIRERPAAIPIEIKSID 406
> dre:100007897 MGC162290; zgc:162290
Length=909
Score = 52.0 bits (123), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/95 (30%), Positives = 48/95 (50%), Gaps = 3/95 (3%)
Query 41 GTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIKSLLRPK 100
D W LG L++E++VGE PF V ++ E+ + + A+ ++ LLR
Sbjct 762 AVDWWGLGVLIYEMMVGESPFPGDDEEEVFDSIVN-DEVRYPRFLSTEAIGIMRRLLRRN 820
Query 101 EEERLGS--RDIKELLKHHFLEGVDCMALHFQPLP 133
E RLGS +D +++ K F +D AL + +P
Sbjct 821 PERRLGSGEKDAEDIKKQPFFRNMDWDALLQRKVP 855
> sce:YFL033C RIM15, TAK1; Glucose-repressible protein kinase
involved in signal transduction during cell proliferation in
response to nutrients, specifically the establishment of stationary
phase; identified as a regulator of IME2; substrate
of Pho80p-Pho85p kinase (EC:2.7.11.1); K12767 serine/threonine-protein
kinase RIM15 [EC:2.7.11.1]
Length=1770
Score = 51.6 bits (122), Expect = 3e-06, Method: Composition-based stats.
Identities = 38/124 (30%), Positives = 58/124 (46%), Gaps = 29/124 (23%)
Query 13 PGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQA 72
P Y APE +++ G+ Q D+ WS+GC+ ELL+G PPF + + AV +
Sbjct 1151 PDYLAPE---TIEGKGEDNKQCDW--------WSVGCIFFELLLGYPPFHAETPDAVFKK 1199
Query 73 LCGYHELSLTQQMP-------------AAAVDFIKSLLRPKEEERLGSRDIKELLKHHFL 119
+ LS Q P A D I+ LL +RLG++ I+E+ H +
Sbjct 1200 I-----LSGVIQWPEFKNEEEEREFLTPEAKDLIEKLLVVDPAKRLGAKGIQEIKDHPYF 1254
Query 120 EGVD 123
+ VD
Sbjct 1255 KNVD 1258
> ath:AT1G48490 kinase
Length=1235
Score = 51.6 bits (122), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 33/118 (27%), Positives = 50/118 (42%), Gaps = 14/118 (11%)
Query 12 VPGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ 71
P Y APE + G G D WS+G +++E LVG PPF + +
Sbjct 1014 TPDYLAPEILLGT------------GHGATADWWSVGIILYEFLVGIPPFNADHPQQIFD 1061
Query 72 ALCGYHEL--SLTQQMPAAAVDFIKSLLRPKEEERLGSRDIKELLKHHFLEGVDCMAL 127
+ + + + M A D I LL +RLG+R E+ +H F + +D L
Sbjct 1062 NILNRNIQWPPVPEDMSHEARDLIDRLLTEDPHQRLGARGAAEVKQHSFFKDIDWNTL 1119
> cel:Y47D3A.16 rsks-1; RSK-pSeventy (RSK-p70 kinase) homolog
family member (rsks-1); K04688 p70 ribosomal S6 kinase [EC:2.7.11.1]
Length=580
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 32/91 (35%), Positives = 48/91 (52%), Gaps = 7/91 (7%)
Query 38 CGYG--TDCWSLGCLVHELLVGEPPFKSVS-STAVAQALCGYHELSLTQQMPAAAVDFIK 94
CG+G D WSLG L+ ++L G PPF + + + + L G L+L + A D IK
Sbjct 260 CGHGKAVDWWSLGALMFDMLTGGPPFTAENRRKTIDKILKG--RLTLPAYLSNEARDLIK 317
Query 95 SLLRPKEEERLGS--RDIKELLKHHFLEGVD 123
LL+ + RLG+ D +E+ H F + D
Sbjct 318 KLLKRHVDTRLGAGLSDAEEIKSHAFFKTTD 348
> cel:H42K12.1 pdk-1; PDK-class protein kinase family member (pdk-1);
K06276 3-phosphoinositide dependent protein kinase-1
[EC:2.7.11.1]
Length=636
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 55/113 (48%), Gaps = 16/113 (14%)
Query 15 YAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALC 74
Y +PE + +DGD G TD W LGC++ + L G+PPF++V+ + + +
Sbjct 276 YVSPE----MLADGD--------VGPQTDIWGLGCILFQCLAGQPPFRAVNQYHLLKRIQ 323
Query 75 GYHELSLTQQMPAAAVDFIKSLLRPKEEERLGSRDIKELLKHHFLEGVDCMAL 127
+ S + P A + I +L R+ S +EL+ H F E VD + +
Sbjct 324 EL-DFSFPEGFPEEASEIIAKILVRDPSTRITS---QELMAHKFFENVDWVNI 372
> ath:AT1G79250 AGC1.7; AGC1.7 (AGC KINASE 1.7); kinase
Length=555
Score = 51.2 bits (121), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 45/90 (50%), Gaps = 4/90 (4%)
Query 37 GCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQ--QMPAAAVDFIK 94
G G D W+ G ++ELL G PFK + A + G L + + +AA D IK
Sbjct 396 GHGSAVDWWTFGIFIYELLYGATPFKGQGNRATLHNVIG-QALRFPEVPHVSSAARDLIK 454
Query 95 SLLRPKEEERLG-SRDIKELLKHHFLEGVD 123
LL + ++R+ R E+ +H F EGV+
Sbjct 455 GLLVKEPQKRIAYKRGATEIKQHPFFEGVN 484
> hsa:5586 PKN2, MGC150606, MGC71074, PAK2, PRK2, PRKCL2, PRO2042,
Pak-2; protein kinase N2 (EC:2.7.11.13); K06071 protein
kinase N [EC:2.7.11.13]
Length=984
Score = 51.2 bits (121), Expect = 4e-06, Method: Composition-based stats.
Identities = 29/89 (32%), Positives = 46/89 (51%), Gaps = 3/89 (3%)
Query 41 GTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIKSLLRPK 100
D W LG L++E+LVGE PF V ++ E+ + + A+ ++ LLR
Sbjct 837 AVDWWGLGVLIYEMLVGESPFPGDDEEEVFDSIVN-DEVRYPRFLSTEAISIMRRLLRRN 895
Query 101 EEERLGS--RDIKELLKHHFLEGVDCMAL 127
E RLG+ +D +++ KH F +D AL
Sbjct 896 PERRLGASEKDAEDVKKHPFFRLIDWSAL 924
> ath:AT1G45160 kinase
Length=1067
Score = 51.2 bits (121), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 38/121 (31%), Positives = 51/121 (42%), Gaps = 14/121 (11%)
Query 12 VPGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAV-A 70
P Y APE + + GY D WS G ++ ELL G PPF + +
Sbjct 858 TPDYLAPEILLGTEH------------GYAADWWSAGIVLFELLTGIPPFTASRPEKIFD 905
Query 71 QALCGYHEL-SLTQQMPAAAVDFIKSLLRPKEEERLGSRDIKELLKHHFLEGVDCMALHF 129
L G + +M A D I LL + E+RLG+ E+ H F +GVD L
Sbjct 906 NILNGKMPWPDVPGEMSYEAQDLINRLLVHEPEKRLGANGAAEVKSHPFFQGVDWENLAL 965
Query 130 Q 130
Q
Sbjct 966 Q 966
> dre:562242 ribosomal protein S6 kinase b, polypeptide 1-like;
K04688 p70 ribosomal S6 kinase [EC:2.7.11.1]
Length=507
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 34/120 (28%), Positives = 55/120 (45%), Gaps = 15/120 (12%)
Query 37 GCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLTQQMPAAAVDFIKSL 96
G D WSLG L++++L G PPF + + + +L+L + A D +K L
Sbjct 244 GHNRAVDWWSLGALMYDMLTGAPPFTAENRKKTIDKILKC-KLNLPPYLTQEARDLLKKL 302
Query 97 LRPKEEERLGS--RDIKELLKHHFLEGV---DCMALHFQPLPL--------DVMQYDFAF 143
L+ R+G+ RD+ ++ +H F + D +A P P DV Q+D F
Sbjct 303 LKRNASVRIGASPRDVLDIQRHSFFRHINWDDLLAFKIDP-PFKPFLQSADDVSQFDSKF 361
> xla:414693 stk38l, MGC83214, ndr2; serine/threonine kinase 38
like (EC:2.7.11.1); K08790 serine/threonine kinase 38 [EC:2.7.11.1]
Length=464
Score = 50.8 bits (120), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 43/149 (28%), Positives = 67/149 (44%), Gaps = 28/149 (18%)
Query 1 RQTAVIYPHAVVPGYAAPEASVSVDSDGDSVAQYDFGCGYGT--DCWSLGCLVHELLVGE 58
RQ A Y P Y APE V GY D WSLG +++E+L+G
Sbjct 277 RQLA--YSTVGTPDYIAPEVFVQ--------------TGYNKLCDWWSLGVIMYEMLIGY 320
Query 59 PPFKSVSSTAVAQALCGYHE-LSLTQQMPAA--AVDFIKSLLR--PKEEERLGSRDIKEL 113
PPF S + + + + E L ++P + + D I LR E R+GS ++E+
Sbjct 321 PPFCSETPQETYRKVMNWKETLVFPPEVPTSEKSKDLI---LRFCTDSENRIGSNGVEEI 377
Query 114 LKHHFLEGVDCMALHFQP--LPLDVMQYD 140
F +GVD + +P +P+++ D
Sbjct 378 KSQPFFDGVDWGHIRERPAAIPIEIKSID 406
> dre:567039 cdc42bpb, si:dkeyp-93h6.2, wu:fb98c02, wu:fc48b05,
wu:fc90d12; CDC42 binding protein kinase beta (DMPK-like)
(EC:2.7.11.1); K08286 protein-serine/threonine kinase [EC:2.7.11.-]
Length=1708
Score = 50.4 bits (119), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 36/116 (31%), Positives = 54/116 (46%), Gaps = 12/116 (10%)
Query 12 VPGYAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQ 71
P Y +PE +++ D + +Y C D WSLG ++E+L GE PF + S
Sbjct 239 TPDYISPEILQAME---DGMGKYGPEC----DWWSLGVCMYEMLYGETPFYAESLVETYG 291
Query 72 ALCGYHEL----SLTQQMPAAAVDFIKSLLRPKEEERLGSRDIKELLKHHFLEGVD 123
+ + E S + A D I+ L+ + E RLG I+E KH F G+D
Sbjct 292 KIMNHEERFQFPSHITDVSEDAKDLIQRLICSR-ERRLGQHGIEEFRKHAFFSGID 346
> ath:AT1G53700 WAG1; WAG1 (WAG 1); kinase/ protein serine/threonine
kinase
Length=476
Score = 50.4 bits (119), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 46/89 (51%), Gaps = 3/89 (3%)
Query 37 GCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELSLT--QQMPAAAVDFIK 94
G G G D W+ G ++E+L G PFK + + + +++ T ++ A D I+
Sbjct 315 GHGSGVDWWAFGIFLYEMLYGTTPFKGGTKEQTLRNIVSNDDVAFTLEEEGMVEAKDLIE 374
Query 95 SLLRPKEEERLG-SRDIKELLKHHFLEGV 122
LL +RLG +R +++ +H F EG+
Sbjct 375 KLLVKDPRKRLGCARGAQDIKRHEFFEGI 403
> dre:558488 serine/threonine-protein kinase MRCK alpha-like;
K08788 dystrophia myotonica-protein kinase [EC:2.7.11.1]
Length=367
Score = 50.4 bits (119), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 42/130 (32%), Positives = 62/130 (47%), Gaps = 21/130 (16%)
Query 12 VPGYAAPEASVSVDSDGDSVAQYDFGCGYGTDC--WSLGCLVHELLVGEPPF--KSVSST 67
P Y +PE +V++ G GYG+DC W+LG +E+L+G PF +S+S T
Sbjct 221 TPDYLSPELLRAVEAGG----------GYGSDCDWWALGVCAYEMLLGTTPFYAESISET 270
Query 68 AVA----QALCGYHELSLTQQMPAAAVDFIKSLLRPKEEERLGSRDIKELLKHHFLEGVD 123
+ G+ + PA A+ + SLL + ERLGS + H F G+D
Sbjct 271 YAKIIHFKDYFGFPASGVQVSEPARAL--VCSLL-CERSERLGSGGSADFRSHPFFSGLD 327
Query 124 CMALHFQPLP 133
+LH P P
Sbjct 328 WSSLHLHPPP 337
> hsa:9252 RPS6KA5, MGC1911, MSK1, MSPK1, RLPK; ribosomal protein
S6 kinase, 90kDa, polypeptide 5 (EC:2.7.11.1); K04445 mitogen-,stress
activated protein kinases [EC:2.7.11.1]
Length=802
Score = 50.4 bits (119), Expect = 6e-06, Method: Composition-based stats.
Identities = 38/111 (34%), Positives = 53/111 (47%), Gaps = 7/111 (6%)
Query 29 DSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYH----ELSLTQQ 84
D V D G D WSLG L++ELL G PF +V +QA E Q+
Sbjct 223 DIVRGGDSGHDKAVDWWSLGVLMYELLTGASPF-TVDGEKNSQAEISRRILKSEPPYPQE 281
Query 85 MPAAAVDFIKSLLRPKEEERL--GSRDIKELLKHHFLEGVDCMALHFQPLP 133
M A A D I+ LL ++RL G RD E+ +H F + ++ L + +P
Sbjct 282 MSALAKDLIQRLLMKDPKKRLGCGPRDADEIKEHLFFQKINWDDLAAKKVP 332
Score = 33.5 bits (75), Expect = 0.88, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 28/49 (57%), Gaps = 12/49 (24%)
Query 15 YAAPEASVSVDSDGDSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKS 63
YAAPE ++ +G YD C D WSLG +++ +L G+ PF+S
Sbjct 588 YAAPEL---LNQNG-----YDESC----DLWSLGVILYTMLSGQVPFQS 624
> mmu:73086 Rps6ka5, 3110005L17Rik, 6330404E13Rik, AI854034, MGC28385,
MSK1, MSPK1, RLPK; ribosomal protein S6 kinase, polypeptide
5 (EC:2.7.11.1); K04445 mitogen-,stress activated protein
kinases [EC:2.7.11.1]
Length=863
Score = 50.4 bits (119), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 37/111 (33%), Positives = 52/111 (46%), Gaps = 7/111 (6%)
Query 29 DSVAQYDFGCGYGTDCWSLGCLVHELLVGEPPFKSVSSTAVAQALCGYHELS----LTQQ 84
D V D G D WSLG L++ELL G PF +V +QA L Q+
Sbjct 222 DIVRGGDSGHDKAVDWWSLGVLMYELLTGASPF-TVDGEKNSQAEISRRILKSEPPYPQE 280
Query 85 MPAAAVDFIKSLLRPKEEERL--GSRDIKELLKHHFLEGVDCMALHFQPLP 133
M A D ++ LL ++RL G RD +E+ +H F E + L + +P
Sbjct 281 MSTVAKDLLQRLLMKDPKKRLGCGPRDAEEIKEHLFFEKIKWDDLAAKKVP 331
Lambda K H
0.318 0.133 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9954723792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40