bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2496_orf2
Length=182
Score E
Sequences producing significant alignments: (Bits) Value
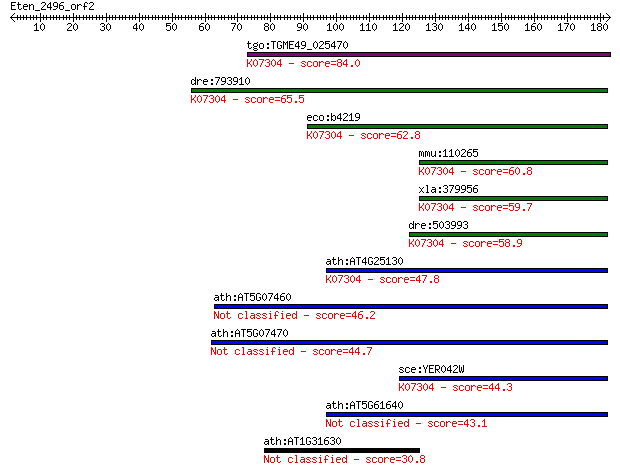
tgo:TGME49_025470 peptide methionine sulfoxide reductase, puta... 84.0 3e-16
dre:793910 si:dkey-221h15.1 (EC:1.8.4.11); K07304 peptide-meth... 65.5 1e-10
eco:b4219 msrA, ECK4215, JW4178, pms, pmsR; methionine sulfoxi... 62.8 7e-10
mmu:110265 Msra, 2310045J23Rik, 6530413P12Rik, MGC101976, MSR-... 60.8 3e-09
xla:379956 msra.1, MGC64457, msra; methionine sulfoxide reduct... 59.7 6e-09
dre:503993 im:7149628; K07304 peptide-methionine (S)-S-oxide r... 58.9 9e-09
ath:AT4G25130 peptide methionine sulfoxide reductase, putative... 47.8 2e-05
ath:AT5G07460 PMSR2; PMSR2 (PEPTIDEMETHIONINE SULFOXIDE REDUCT... 46.2 6e-05
ath:AT5G07470 PMSR3; PMSR3 (PEPTIDEMETHIONINE SULFOXIDE REDUCT... 44.7 2e-04
sce:YER042W MXR1; Methionine-S-sulfoxide reductase, involved i... 44.3 2e-04
ath:AT5G61640 PMSR1; PMSR1 (PEPTIDEMETHIONINE SULFOXIDE REDUCT... 43.1 5e-04
ath:AT1G31630 AGL86; AGL86 (AGAMOUS-LIKE 86); DNA binding / tr... 30.8 2.5
> tgo:TGME49_025470 peptide methionine sulfoxide reductase, putative
(EC:1.8.4.11); K07304 peptide-methionine (S)-S-oxide
reductase [EC:1.8.4.11]
Length=269
Score = 84.0 bits (206), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 43/110 (39%), Positives = 57/110 (51%), Gaps = 3/110 (2%)
Query 73 VGSSPAAAAAAAPAAAAAAGAPPAGAAAAPAAAPAAAAAAAPPAAAAAAESIVLGGGCFW 132
V SS + + PA+ A+ A P A ++LG GCFW
Sbjct 59 VDSSLQMSRLSPPASCASTSAGDDRGTQLEQHTPNFLPGTD---RTAPFSRVLLGAGCFW 115
Query 133 GLEKLFVEEFKDKLEATCVGYAGGHTPQPTYAQVCSSKTGHFEALKISFF 182
G+EKLF +EF +L AT VGYAGG PTY +VC+ TGH+E ++I FF
Sbjct 116 GVEKLFRKEFGSQLRATTVGYAGGAKDHPTYKEVCTGNTGHYEVVEIQFF 165
> dre:793910 si:dkey-221h15.1 (EC:1.8.4.11); K07304 peptide-methionine
(S)-S-oxide reductase [EC:1.8.4.11]
Length=235
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 63/128 (49%), Gaps = 9/128 (7%)
Query 56 LIAFTSMSLLNSSSGLS-VGSSPAAAAAAAPAAAAAAGAPPAGAAAAPAAAPAAAAAAAP 114
++A TS+ L+ S +G + +P A P + +A
Sbjct 1 MVARTSVRLIWRQFIQSRMGEMSSKVQMISPEEAL----PGREQSIKVSAKHDVNGNRTV 56
Query 115 PAAAAAAESIVLGGGCFWGLEKLFVEEFKDK-LEATCVGYAGGHTPQPTYAQVCSSKTGH 173
P + ++ G GCFWG E+ F ++ K + +T VGY+GG+TP PTY +VC+ KTGH
Sbjct 57 PPFPEGLQMVLFGMGCFWGAERKF---WRQKGVYSTQVGYSGGYTPNPTYEEVCTGKTGH 113
Query 174 FEALKISF 181
E +++ F
Sbjct 114 TEVVRVVF 121
> eco:b4219 msrA, ECK4215, JW4178, pms, pmsR; methionine sulfoxide
reductase A (EC:1.8.4.11); K07304 peptide-methionine (S)-S-oxide
reductase [EC:1.8.4.11]
Length=212
Score = 62.8 bits (151), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 34/91 (37%), Positives = 46/91 (50%), Gaps = 2/91 (2%)
Query 91 AGAPPAGAAAAPAAAPAAAAAAAPPAAAAAAESIVLGGGCFWGLEKLFVEEFKDKLEATC 150
A A P P A A + E + GCFWG+E+LF + + +T
Sbjct 13 ADALPGRNTPMPVATLHAVNGHSMTNVPDGMEIAIFAMGCFWGVERLFWQ--LPGVYSTA 70
Query 151 VGYAGGHTPQPTYAQVCSSKTGHFEALKISF 181
GY GG+TP PTY +VCS TGH EA++I +
Sbjct 71 AGYTGGYTPNPTYREVCSGDTGHAEAVRIVY 101
> mmu:110265 Msra, 2310045J23Rik, 6530413P12Rik, MGC101976, MSR-A;
methionine sulfoxide reductase A (EC:1.8.4.11); K07304
peptide-methionine (S)-S-oxide reductase [EC:1.8.4.11]
Length=233
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 28/57 (49%), Positives = 37/57 (64%), Gaps = 2/57 (3%)
Query 125 VLGGGCFWGLEKLFVEEFKDKLEATCVGYAGGHTPQPTYAQVCSSKTGHFEALKISF 181
V G GCFWG E+ F + +T VG+AGGHT PTY +VCS KTGH E +++ +
Sbjct 67 VFGMGCFWGAERKFW--VLKGVYSTQVGFAGGHTRNPTYKEVCSEKTGHAEVVRVVY 121
> xla:379956 msra.1, MGC64457, msra; methionine sulfoxide reductase
A, gene 1 (EC:1.8.4.11); K07304 peptide-methionine (S)-S-oxide
reductase [EC:1.8.4.11]
Length=211
Score = 59.7 bits (143), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 26/58 (44%), Positives = 40/58 (68%), Gaps = 4/58 (6%)
Query 125 VLGGGCFWGLEKLFVEEFKDK-LEATCVGYAGGHTPQPTYAQVCSSKTGHFEALKISF 181
+ G GCFWG E+ F ++ K + +T VGY+GG+TP P Y +VCS +TGH E +++ +
Sbjct 46 IFGMGCFWGAERKF---WRQKGVYSTQVGYSGGYTPNPLYEEVCSGRTGHAEVVRVVY 100
> dre:503993 im:7149628; K07304 peptide-methionine (S)-S-oxide
reductase [EC:1.8.4.11]
Length=160
Score = 58.9 bits (141), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 29/60 (48%), Positives = 38/60 (63%), Gaps = 2/60 (3%)
Query 122 ESIVLGGGCFWGLEKLFVEEFKDKLEATCVGYAGGHTPQPTYAQVCSSKTGHFEALKISF 181
E I+ G GCFWG E+ F + + +T VGYAGG T PTY +VCS TGH E +++ F
Sbjct 81 EMIMFGMGCFWGAERRFWK--ITGVFSTQVGYAGGFTSNPTYHEVCSGLTGHTEVVRVVF 138
> ath:AT4G25130 peptide methionine sulfoxide reductase, putative;
K07304 peptide-methionine (S)-S-oxide reductase [EC:1.8.4.11]
Length=258
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 44/88 (50%), Gaps = 5/88 (5%)
Query 97 GAAAAPAAAPAAAAAAAPPAA---AAAAESIVLGGGCFWGLEKLFVEEFKDKLEATCVGY 153
G+ A P++AA A P ++ + G GCFWG+E + + + T VGY
Sbjct 64 GSRPQAQADPSSAAIAQGPDDDVPSSGQQFAQFGAGCFWGVELAY--QRVPGVTKTEVGY 121
Query 154 AGGHTPQPTYAQVCSSKTGHFEALKISF 181
+ G P+Y VC+ TGH E +++ +
Sbjct 122 SHGIVHNPSYEDVCTGTTGHNEVVRVQY 149
> ath:AT5G07460 PMSR2; PMSR2 (PEPTIDEMETHIONINE SULFOXIDE REDUCTASE
2); oxidoreductase, acting on sulfur group of donors,
disulfide as acceptor / peptide-methionine-(S)-S-oxide reductase
Length=218
Score = 46.2 bits (108), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 32/119 (26%), Positives = 47/119 (39%), Gaps = 12/119 (10%)
Query 63 SLLNSSSGLSVGSSPAAAAAAAPAAAAAAGAPPAGAAAAPAAAPAAAAAAAPPAAAAAAE 122
S L + V +SP+ A P A P+ A P A E
Sbjct 3 SSLKTQEPQVVETSPSPVAQEPPQVADKPAIVPSPIAQEPDND----------VPAPGNE 52
Query 123 SIVLGGGCFWGLEKLFVEEFKDKLEATCVGYAGGHTPQPTYAQVCSSKTGHFEALKISF 181
GCFWG+E F + + T VGY G + P+Y VC++ T H E +++ +
Sbjct 53 FAEFAAGCFWGVELAF--QRIPGVTVTEVGYTHGISHNPSYEDVCTNTTNHAEVVRVQY 109
> ath:AT5G07470 PMSR3; PMSR3 (PEPTIDEMETHIONINE SULFOXIDE REDUCTASE
3); oxidoreductase, acting on sulfur group of donors,
disulfide as acceptor / peptide-methionine-(S)-S-oxide reductase
Length=202
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/120 (28%), Positives = 46/120 (38%), Gaps = 27/120 (22%)
Query 62 MSLLNSSSGLSVGSSPAAAAAAAPAAAAAAGAPPAGAAAAPAAAPAAAAAAAPPAAAAAA 121
M++LN L +GSS +P A PA P A
Sbjct 1 MNILNR---LGLGSSGQTNMDPSPIAQGNDDDTPA------------------PGNQFAQ 39
Query 122 ESIVLGGGCFWGLEKLFVEEFKDKLEATCVGYAGGHTPQPTYAQVCSSKTGHFEALKISF 181
G GCFWG+E F + + T GY G P+Y VCS TGH E +++ +
Sbjct 40 ----FGAGCFWGVELAF--QRVPGVTQTEAGYTQGTVDNPSYGDVCSGTTGHSEVVRVQY 93
> sce:YER042W MXR1; Methionine-S-sulfoxide reductase, involved
in the response to oxidative stress; protects iron-sulfur clusters
from oxidative inactivation along with MXR2; involved
in the regulation of lifespan (EC:1.8.4.11); K07304 peptide-methionine
(S)-S-oxide reductase [EC:1.8.4.11]
Length=184
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 34/69 (49%), Gaps = 6/69 (8%)
Query 119 AAAESIVLGGGCFWGLEKLFVEEFKDKLEATCVGYAGGHTPQP------TYAQVCSSKTG 172
A + I L GCFWG E ++ + D++ VGYA G + +Y +VC T
Sbjct 14 AKDKLITLACGCFWGTEHMYRKYLNDRIVDCKVGYANGEESKKDSPSSVSYKRVCGGDTD 73
Query 173 HFEALKISF 181
E L++S+
Sbjct 74 FAEVLQVSY 82
> ath:AT5G61640 PMSR1; PMSR1 (PEPTIDEMETHIONINE SULFOXIDE REDUCTASE
1); oxidoreductase, acting on sulfur group of donors,
disulfide as acceptor / peptide-methionine-(S)-S-oxide reductase
Length=202
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 25/86 (29%), Positives = 38/86 (44%), Gaps = 3/86 (3%)
Query 97 GAAAAPAAAPAAAAAAAPPAAAAAAESIV-LGGGCFWGLEKLFVEEFKDKLEATCVGYAG 155
G++ P+ A A A G GCFW +E + + + T VGY+
Sbjct 10 GSSRQTNMDPSPIAQVIDDEAPAPGNQFTQFGAGCFWSVELAY--QRVPGVTQTEVGYSQ 67
Query 156 GHTPQPTYAQVCSSKTGHFEALKISF 181
G T P+Y VCS T H E +++ +
Sbjct 68 GITHDPSYKDVCSGTTNHAEIVRVQY 93
> ath:AT1G31630 AGL86; AGL86 (AGAMOUS-LIKE 86); DNA binding /
transcription factor
Length=339
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 24/47 (51%), Positives = 24/47 (51%), Gaps = 1/47 (2%)
Query 78 AAAAAAAPAAAAAAGAPPAGAAAAPAAAPAAAAAAAPPAAAAAAESI 124
A A AAP A A AGA P A A AAP A A A PP A E I
Sbjct 186 VAGAGAAPLAVAGAGASPL-AVAGVGAAPLAVAGAGPPMAQNQYEPI 231
Lambda K H
0.321 0.131 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4912245712
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40