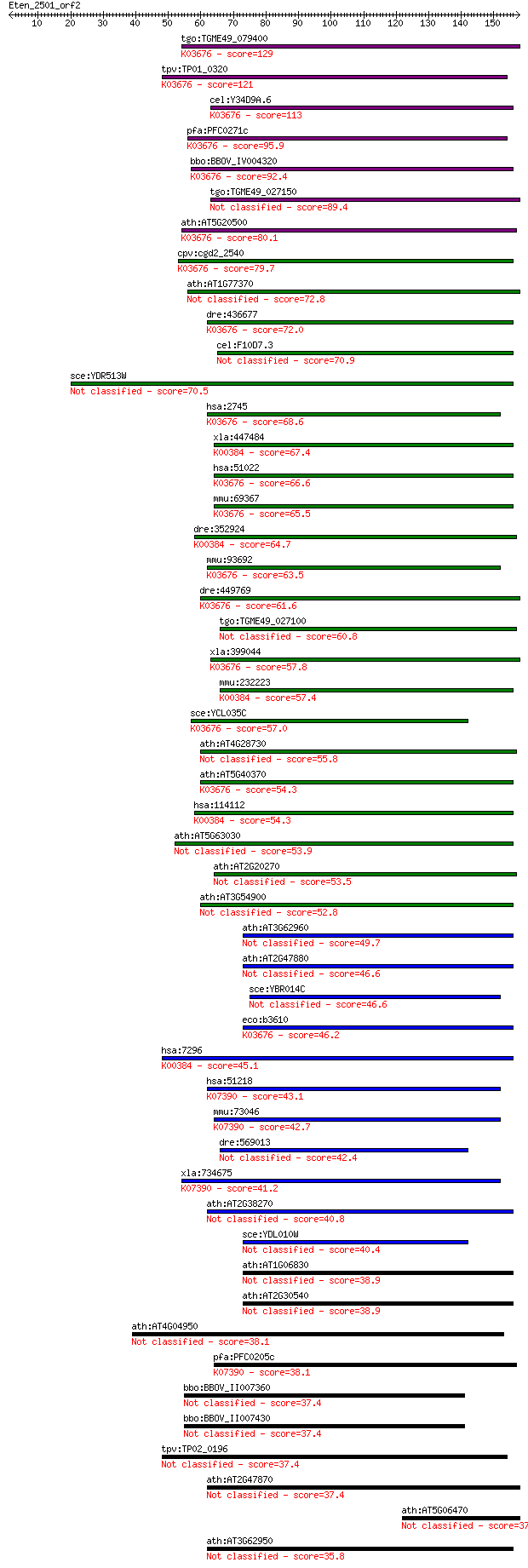
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2501_orf2
Length=157
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_079400 glutaredoxin, putative ; K03676 glutaredoxin 3 129 3e-30
tpv:TP01_0320 glutaredoxin-like protein; K03676 glutaredoxin 3 121 8e-28
cel:Y34D9A.6 glrx-10; GLutaRedoXin family member (glrx-10); K0... 113 3e-25
pfa:PFC0271c GRX1, GRX-1; glutaredoxin 1; K03676 glutaredoxin 3 95.9
bbo:BBOV_IV004320 23.m06161; glutaredoxin-like protein; K03676... 92.4 5e-19
tgo:TGME49_027150 glutaredoxin, putative 89.4 4e-18
ath:AT5G20500 glutaredoxin, putative; K03676 glutaredoxin 3 80.1
cpv:cgd2_2540 glutaredoxin related protein ; K03676 glutaredox... 79.7 4e-15
ath:AT1G77370 glutaredoxin, putative 72.8 4e-13
dre:436677 glrx2, zgc:92698; glutaredoxin 2; K03676 glutaredox... 72.0 7e-13
cel:F10D7.3 hypothetical protein 70.9 2e-12
sce:YDR513W GRX2, TTR1; Cytoplasmic glutaredoxin, thioltransfe... 70.5 2e-12
hsa:2745 GLRX, GRX, GRX1, MGC117407; glutaredoxin (thioltransf... 68.6 8e-12
xla:447484 txnrd3, MGC81848; thioredoxin reductase 3 (EC:1.8.1... 67.4 2e-11
hsa:51022 GLRX2, GRX2, bA101E13.1; glutaredoxin 2; K03676 glut... 66.6 3e-11
mmu:69367 Glrx2, 1700010P22Rik, AI645710, Grx2; glutaredoxin 2... 65.5 8e-11
dre:352924 txnrd1, TrxR1, cb682, fb83a08, wu:fb83a08; thioredo... 64.7 1e-10
mmu:93692 Glrx, C86710, D13Wsu156e, Glrx1, Grx1, TTase; glutar... 63.5 3e-10
dre:449769 glrx, MGC113837, grx, wu:fc38f02, zgc:103707; gluta... 61.6 1e-09
tgo:TGME49_027100 glutaredoxin, putative 60.8 2e-09
xla:399044 glrx, MGC68461, grx, grx1; glutaredoxin (thioltrans... 57.8 1e-08
mmu:232223 Txnrd3, AI196535, TR2, Tgr; thioredoxin reductase 3... 57.4 2e-08
sce:YCL035C GRX1; Grx1p; K03676 glutaredoxin 3 57.0
ath:AT4G28730 glutaredoxin family protein 55.8 6e-08
ath:AT5G40370 glutaredoxin, putative; K03676 glutaredoxin 3 54.3
hsa:114112 TXNRD3, TGR, TR2, TRXR3; thioredoxin reductase 3 (E... 54.3 2e-07
ath:AT5G63030 glutaredoxin, putative 53.9 2e-07
ath:AT2G20270 glutaredoxin family protein 53.5 3e-07
ath:AT3G54900 CXIP1; CXIP1 (CAX INTERACTING PROTEIN 1); antipo... 52.8 4e-07
ath:AT3G62960 glutaredoxin family protein 49.7 3e-06
ath:AT2G47880 glutaredoxin family protein 46.6 3e-05
sce:YBR014C GRX7; Cis-golgi localized monothiol glutaredoxin; ... 46.6 3e-05
eco:b3610 grxC, ECK3600, JW3585, yibM; glutaredoxin 3; K03676 ... 46.2 5e-05
hsa:7296 TXNRD1, GRIM-12, MGC9145, TR, TR1, TRXR1, TXNR; thior... 45.1 1e-04
hsa:51218 GLRX5, C14orf87, FLB4739, GRX5, MGC14129, PR01238, P... 43.1 4e-04
mmu:73046 Glrx5, 2310004O13Rik, 2900070E19Rik, AU020725; gluta... 42.7 4e-04
dre:569013 MGC152951; zgc:152951 42.4 6e-04
xla:734675 glrx5, MGC116473, grx5; glutaredoxin 5; K07390 mono... 41.2 0.001
ath:AT2G38270 CXIP2; CXIP2 (CAX-INTERACTING PROTEIN 2); electr... 40.8 0.002
sce:YDL010W GRX6; Cis-golgi localized monothiol glutaredoxin t... 40.4 0.002
ath:AT1G06830 glutaredoxin family protein 38.9 0.006
ath:AT2G30540 glutaredoxin family protein 38.9 0.007
ath:AT4G04950 thioredoxin family protein 38.1 0.011
pfa:PFC0205c PfGLP-1; 1-cys-glutaredoxin-like protein-1; K0739... 38.1 0.013
bbo:BBOV_II007360 18.m06610; Thioredoxin-like protein 2 37.4
bbo:BBOV_II007430 18.m06616; thioredoxin-like protein 2 37.4
tpv:TP02_0196 hypothetical protein 37.4 0.021
ath:AT2G47870 glutaredoxin family protein 37.4 0.022
ath:AT5G06470 glutaredoxin family protein 37.0 0.027
ath:AT3G62950 glutaredoxin family protein 35.8 0.053
> tgo:TGME49_079400 glutaredoxin, putative ; K03676 glutaredoxin
3
Length=112
Score = 129 bits (325), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 57/105 (54%), Positives = 77/105 (73%), Gaps = 1/105 (0%)
Query 54 LNSAQDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAI 113
L S V +W D L+ HKV +F+K++CPYC+ AI A +S+ DMHVE+IE +P+ +AI
Sbjct 5 LASEAHVAEWADRLIKQHKVVVFSKSNCPYCRKAIEAFQSVKAKDMHVEEIEGSPYMDAI 64
Query 114 QDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTL-QKLIAA 157
QDY+K++TGARSVPRVFI G F GG +DT+ +GTL +KL AA
Sbjct 65 QDYMKQQTGARSVPRVFIGGQFLGGAEDTIRAKADGTLVEKLRAA 109
> tpv:TP01_0320 glutaredoxin-like protein; K03676 glutaredoxin
3
Length=151
Score = 121 bits (304), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 51/107 (47%), Positives = 77/107 (71%), Gaps = 1/107 (0%)
Query 48 FASTMALNSAQDVPQ-WVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIEN 106
++ T+ A+ P+ WVDSLV HKV +F+K++CPYC A AL+ LN+ D+HVE++++
Sbjct 38 YSFTLPFKMAEKTPKDWVDSLVKKHKVVVFSKSYCPYCTRAKDALKKLNLHDLHVEELDS 97
Query 107 NPHCNAIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQK 153
NP+ + +QDYL + TGARSVPRVF+NG F+G TV+ V++G +
Sbjct 98 NPNMDQVQDYLNQLTGARSVPRVFVNGRFYGDSTKTVSDVESGKFME 144
> cel:Y34D9A.6 glrx-10; GLutaRedoXin family member (glrx-10);
K03676 glutaredoxin 3
Length=105
Score = 113 bits (282), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 54/95 (56%), Positives = 66/95 (69%), Gaps = 2/95 (2%)
Query 63 WVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVE--QIENNPHCNAIQDYLKEK 120
+VD L+ KV +F+K++CPYC A AAL S+NV ++ +I+ CN IQDYL
Sbjct 5 FVDGLLQSSKVVVFSKSYCPYCHKARAALESVNVKPDALQWIEIDERKDCNEIQDYLGSL 64
Query 121 TGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
TGARSVPRVFING FFGGGDDT AG KNG L L+
Sbjct 65 TGARSVPRVFINGKFFGGGDDTAAGAKNGKLAALL 99
> pfa:PFC0271c GRX1, GRX-1; glutaredoxin 1; K03676 glutaredoxin
3
Length=111
Score = 95.9 bits (237), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 46/99 (46%), Positives = 62/99 (62%), Gaps = 1/99 (1%)
Query 56 SAQDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNV-SDMHVEQIENNPHCNAIQ 114
+++ V +WV+ ++ + + +FAKT CPYC AI+ L+ N+ S MHVE IE NP IQ
Sbjct 4 TSEAVKKWVNKIIEENIIAVFAKTECPYCIKAISILKGYNLNSHMHVENIEKNPDMANIQ 63
Query 115 DYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQK 153
YLKE TG SVPR+FIN GG DD V G L++
Sbjct 64 AYLKELTGKSSVPRIFINKDVVGGCDDLVKENDEGKLKE 102
> bbo:BBOV_IV004320 23.m06161; glutaredoxin-like protein; K03676
glutaredoxin 3
Length=129
Score = 92.4 bits (228), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 42/99 (42%), Positives = 62/99 (62%), Gaps = 0/99 (0%)
Query 57 AQDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDY 116
D+ WV+ +N KV +F+KT CPYC A L S+ +D+ + Q+++NP I +Y
Sbjct 25 GSDISHWVNESINKSKVVVFSKTTCPYCIKANGILNSVAPNDLTIIQLDDNPDRAEIMEY 84
Query 117 LKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
+E TGA +VPRVFI G FFG TVA ++G L+K++
Sbjct 85 FRETTGAATVPRVFIGGKFFGDCSKTVAANESGELKKVL 123
> tgo:TGME49_027150 glutaredoxin, putative
Length=332
Score = 89.4 bits (220), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 40/95 (42%), Positives = 59/95 (62%), Gaps = 0/95 (0%)
Query 63 WVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTG 122
W+ + HKV +FA ++CPYC A+ LR+ V D+ I+ + IQD L+E TG
Sbjct 234 WIKEKIAKHKVVVFAMSYCPYCDTALEILRNAGVKDLGDVMIDRMDYTPQIQDILEEMTG 293
Query 123 ARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIAA 157
AR+VPRVFI+G FFGG D +G L+++++A
Sbjct 294 ARTVPRVFIDGIFFGGCSDLEEAEASGKLKEILSA 328
> ath:AT5G20500 glutaredoxin, putative; K03676 glutaredoxin 3
Length=135
Score = 80.1 bits (196), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 40/105 (38%), Positives = 63/105 (60%), Gaps = 3/105 (2%)
Query 54 LNSAQDVPQ--WVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCN 111
++SA P+ +V ++ HK+ IF+K++CPYC+ A + R L+ +V +++
Sbjct 23 VSSAASSPEADFVKKTISSHKIVIFSKSYCPYCKKAKSVFRELDQVP-YVVELDEREDGW 81
Query 112 AIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIA 156
+IQ L E G R+VP+VFING GG DDTV ++G L KL+
Sbjct 82 SIQTALGEIVGRRTVPQVFINGKHLGGSDDTVDAYESGELAKLLG 126
> cpv:cgd2_2540 glutaredoxin related protein ; K03676 glutaredoxin
3
Length=108
Score = 79.7 bits (195), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 39/103 (37%), Positives = 63/103 (61%), Gaps = 4/103 (3%)
Query 53 ALNSAQDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNA 112
A+NS + V+S ++ + + +K++CPYC AI +L+S S + V QI+
Sbjct 7 AMNS---IKLLVESFISSGDICVISKSYCPYCIKAINSLKSAGYSPL-VMQIDGRVDTKE 62
Query 113 IQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
IQDY +E TG+ +VPRVF+ G F GG DDT+ +++G+L +
Sbjct 63 IQDYCRELTGSGTVPRVFVKGRFIGGCDDTLKLLEDGSLSSFV 105
> ath:AT1G77370 glutaredoxin, putative
Length=130
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 58/102 (56%), Gaps = 1/102 (0%)
Query 56 SAQDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQD 115
+A V +V + + +K+ IF+K++CPYC + L VE ++ + IQ
Sbjct 29 AANSVSAFVQNAILSNKIVIFSKSYCPYCLRSKRIFSQLKEEPFVVE-LDQREDGDQIQY 87
Query 116 YLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIAA 157
L E G R+VP+VF+NG GG DD A +++G LQKL+AA
Sbjct 88 ELLEFVGRRTVPQVFVNGKHIGGSDDLGAALESGQLQKLLAA 129
> dre:436677 glrx2, zgc:92698; glutaredoxin 2; K03676 glutaredoxin
3
Length=134
Score = 72.0 bits (175), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 41/94 (43%), Positives = 54/94 (57%), Gaps = 1/94 (1%)
Query 62 QWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKT 121
Q+V +V+ + V IF+KT CPYC+ A + + VE E+N +Q+ L E T
Sbjct 18 QFVQDIVSSNCVVIFSKTTCPYCKMAKGVFNEIGATYKVVELDEHNDG-RRLQETLAELT 76
Query 122 GARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
GAR+VPRVFING GGG DT + G L LI
Sbjct 77 GARTVPRVFINGQCIGGGSDTKQLHQQGKLLPLI 110
> cel:F10D7.3 hypothetical protein
Length=146
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 32/94 (34%), Positives = 58/94 (61%), Gaps = 3/94 (3%)
Query 65 DSLVNG---HKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKT 121
D +VN HKV +++KT+CP+ + A L + + DM + +++ + +Q+ LK+ +
Sbjct 35 DKIVNDVMTHKVMVYSKTYCPWSKRLKAILANYEIDDMKIVELDRSNQTEEMQEILKKYS 94
Query 122 GARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
G +VP++FI+G F GG D+T A + G L+ L+
Sbjct 95 GRTTVPQLFISGKFVGGHDETKAIEEKGELRPLL 128
> sce:YDR513W GRX2, TTR1; Cytoplasmic glutaredoxin, thioltransferase,
glutathione-dependent disulfide oxidoreductase involved
in maintaining redox state of target proteins, also exhibits
glutathione peroxidase activity, expression induced in
response to stress
Length=143
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 47/139 (33%), Positives = 73/139 (52%), Gaps = 14/139 (10%)
Query 20 FINLLYPAAFAGRIWARARLRYNSCRTLFASTMALNSAQDVPQWVDSLVNGHKVTIFAKT 79
I ++ FA RI A+ F ST + S + V V L+ +V + AKT
Sbjct 11 LIVIIIITLFATRIIAKR----------FLSTPKMVSQETVAH-VKDLIGQKEVFVAAKT 59
Query 80 HCPYCQAAIAAL-RSLNV--SDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFINGTFF 136
+CPYC+A ++ L + LNV S V +++ + + IQD L+E +G ++VP V+ING
Sbjct 60 YCPYCKATLSTLFQELNVPKSKALVLELDEMSNGSEIQDALEEISGQKTVPNVYINGKHI 119
Query 137 GGGDDTVAGVKNGTLQKLI 155
GG D KNG L +++
Sbjct 120 GGNSDLETLKKNGKLAEIL 138
> hsa:2745 GLRX, GRX, GRX1, MGC117407; glutaredoxin (thioltransferase)
(EC:1.8.4.1); K03676 glutaredoxin 3
Length=106
Score = 68.6 bits (166), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 37/92 (40%), Positives = 51/92 (55%), Gaps = 2/92 (2%)
Query 62 QWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVE--QIENNPHCNAIQDYLKE 119
++V+ + KV +F K CPYC+ A L L + +E I H N IQDYL++
Sbjct 4 EFVNCKIQPGKVVVFIKPTCPYCRRAQEILSQLPIKQGLLEFVDITATNHTNEIQDYLQQ 63
Query 120 KTGARSVPRVFINGTFFGGGDDTVAGVKNGTL 151
TGAR+VPRVFI GG D V+ ++G L
Sbjct 64 LTGARTVPRVFIGKDCIGGCSDLVSLQQSGEL 95
> xla:447484 txnrd3, MGC81848; thioredoxin reductase 3 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=596
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/92 (35%), Positives = 55/92 (59%), Gaps = 1/92 (1%)
Query 64 VDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGA 123
V L++ ++V +F+K+ CPYC SL ++ H +++ + IQ+ L+E TG
Sbjct 13 VKELIDSNRVMVFSKSFCPYCDRVKDLFSSLG-AEYHSLELDECDDGSDIQEALQELTGQ 71
Query 124 RSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
++VP VF+N T GG D T+ K+G+L KL+
Sbjct 72 KTVPNVFVNKTHVGGCDKTLQAHKDGSLAKLL 103
> hsa:51022 GLRX2, GRX2, bA101E13.1; glutaredoxin 2; K03676 glutaredoxin
3
Length=165
Score = 66.6 bits (161), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 51/92 (55%), Gaps = 1/92 (1%)
Query 64 VDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGA 123
+ ++ + V IF+KT C YC A +NV + V +++ + N QD L + TG
Sbjct 61 IQETISDNCVVIFSKTSCSYCTMAKKLFHDMNV-NYKVVELDLLEYGNQFQDALYKMTGE 119
Query 124 RSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
R+VPR+F+NGTF GG DT K G L L+
Sbjct 120 RTVPRIFVNGTFIGGATDTHRLHKEGKLLPLV 151
> mmu:69367 Glrx2, 1700010P22Rik, AI645710, Grx2; glutaredoxin
2 (thioltransferase); K03676 glutaredoxin 3
Length=156
Score = 65.5 bits (158), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 50/92 (54%), Gaps = 1/92 (1%)
Query 64 VDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGA 123
+ ++ + V IF+KT C YC A +NV+ VE ++ + N QD L + TG
Sbjct 53 IQETISNNCVVIFSKTSCSYCSMAKKIFHDMNVNYKAVE-LDMLEYGNQFQDALHKMTGE 111
Query 124 RSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
R+VPR+F+NG F GG DT K G L L+
Sbjct 112 RTVPRIFVNGRFIGGAADTHRLHKEGKLLPLV 143
> dre:352924 txnrd1, TrxR1, cb682, fb83a08, wu:fb83a08; thioredoxin
reductase 1 (EC:1.8.1.9); K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=602
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 33/101 (32%), Positives = 56/101 (55%), Gaps = 5/101 (4%)
Query 58 QDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNV--SDMHVEQIENNPHCNAIQD 115
+ + + L++ V +F+K+ CP+C + LNV + + ++ +E+ + QD
Sbjct 11 EQIRSKIKELIDSSAVVVFSKSFCPFCVKVKDLFKELNVKYNTIELDLMEDGTN---YQD 67
Query 116 YLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIA 156
L E TG ++VP VFIN GG D+T+ K+G LQKL+
Sbjct 68 LLHEMTGQKTVPNVFINKKHIGGCDNTMKAHKDGVLQKLLG 108
> mmu:93692 Glrx, C86710, D13Wsu156e, Glrx1, Grx1, TTase; glutaredoxin
(EC:1.8.4.1); K03676 glutaredoxin 3
Length=107
Score = 63.5 bits (153), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 34/92 (36%), Positives = 49/92 (53%), Gaps = 2/92 (2%)
Query 62 QWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVE--QIENNPHCNAIQDYLKE 119
++V+ + KV +F K CPYC+ L L +E I + +AIQDYL++
Sbjct 4 EFVNCKIQSGKVVVFIKPTCPYCRKTQEILSQLPFKQGLLEFVDITATNNTSAIQDYLQQ 63
Query 120 KTGARSVPRVFINGTFFGGGDDTVAGVKNGTL 151
TGAR+VPRVFI GG D ++ + G L
Sbjct 64 LTGARTVPRVFIGKDCIGGCSDLISMQQTGEL 95
> dre:449769 glrx, MGC113837, grx, wu:fc38f02, zgc:103707; glutaredoxin
(thioltransferase) (EC:1.8.4.1); K03676 glutaredoxin
3
Length=105
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/100 (34%), Positives = 52/100 (52%), Gaps = 2/100 (2%)
Query 60 VPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCN--AIQDYL 117
+ ++V + + K +F K C YC A L H E I+ + + +IQDYL
Sbjct 1 MAEFVKAQIKDGKGVVFCKPTCSYCILAKDVLSKYKFKAGHFELIDISARADMGSIQDYL 60
Query 118 KEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIAA 157
++ TGAR+VPRVFI GGG D ++G L+ ++ A
Sbjct 61 QQITGARTVPRVFIGEDCVGGGSDVEGLDRSGKLEGMLQA 100
> tgo:TGME49_027100 glutaredoxin, putative
Length=112
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/91 (34%), Positives = 49/91 (53%), Gaps = 4/91 (4%)
Query 66 SLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARS 125
S++ HKV +F K H P + L+ +N D+H+ + + C + LKEKTG R
Sbjct 17 SVIKSHKVVVFTKEHDPLSFEVVETLKQINPKDLHIVNL-DKCGCTQAEGLLKEKTGDRP 75
Query 126 VPRVFINGTFFGGGDDTVAGVKNGTLQKLIA 156
PRVFI+G F DD +K+ L++++
Sbjct 76 GPRVFIDGKFV---DDLETSLKDMDLREVLE 103
> xla:399044 glrx, MGC68461, grx, grx1; glutaredoxin (thioltransferase)
(EC:1.8.4.1); K03676 glutaredoxin 3
Length=107
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 50/97 (51%), Gaps = 2/97 (2%)
Query 63 WVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNP--HCNAIQDYLKEK 120
+V S V KVT+F K+ CP+C A L + H+E I+ + +++Q Y +
Sbjct 5 FVQSKVKPSKVTMFEKSSCPFCVRAKGILTKYKFKEGHLEIIDISKLDFMSSLQQYFMQT 64
Query 121 TGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIAA 157
TG +VPR++I GG D V +G L+K + +
Sbjct 65 TGESTVPRIYIGEKCIGGCSDLVPLENSGELEKALES 101
> mmu:232223 Txnrd3, AI196535, TR2, Tgr; thioredoxin reductase
3 (EC:1.8.1.9); K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=652
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/92 (31%), Positives = 53/92 (57%), Gaps = 5/92 (5%)
Query 66 SLVNGHKVTIFAKTHCPYCQAAIAALRSLNV--SDMHVEQIENNPHCNAIQDYLKEKTGA 123
L+ G++V IF+K++CP+ SL V + + ++Q+++ ++Q+ L E +
Sbjct 70 DLIEGNRVMIFSKSYCPHSTRVKELFSSLGVVYNILELDQVDDGA---SVQEVLTEISNQ 126
Query 124 RSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
++VP +F+N GG D T +NG LQKL+
Sbjct 127 KTVPNIFVNKVHVGGCDRTFQAHQNGLLQKLL 158
> sce:YCL035C GRX1; Grx1p; K03676 glutaredoxin 3
Length=110
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/88 (36%), Positives = 49/88 (55%), Gaps = 3/88 (3%)
Query 57 AQDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAAL-RSLNV--SDMHVEQIENNPHCNAI 113
+Q+ + V L+ +++ + +KT+CPYC AA+ L L V S + V Q+ + I
Sbjct 3 SQETIKHVKDLIAENEIFVASKTYCPYCHAALNTLFEKLKVPRSKVLVLQLNDMKEGADI 62
Query 114 QDYLKEKTGARSVPRVFINGTFFGGGDD 141
Q L E G R+VP ++ING GG DD
Sbjct 63 QAALYEINGQRTVPNIYINGKHIGGNDD 90
> ath:AT4G28730 glutaredoxin family protein
Length=174
Score = 55.8 bits (133), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 47/97 (48%), Gaps = 0/97 (0%)
Query 60 VPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKE 119
+ + + V + V I++KT C YC + L V + VE + P +Q L+
Sbjct 69 MEESIRKTVTENTVVIYSKTWCSYCTEVKTLFKRLGVQPLVVELDQLGPQGPQLQKVLER 128
Query 120 KTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIA 156
TG +VP VF+ G GG DTV + G L+ ++A
Sbjct 129 LTGQHTVPNVFVCGKHIGGCTDTVKLNRKGDLELMLA 165
> ath:AT5G40370 glutaredoxin, putative; K03676 glutaredoxin 3
Length=136
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 49/97 (50%), Gaps = 2/97 (2%)
Query 60 VPQW-VDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLK 118
V QW + S + G+ I +KT+CPYC L+ L VE ++ + IQ L
Sbjct 26 VYQWMILSYLIGNFSGICSKTYCPYCVRVKELLQQLGAKFKAVE-LDTESDGSQIQSGLA 84
Query 119 EKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
E TG R+VP VFI G GG D T K+G L L+
Sbjct 85 EWTGQRTVPNVFIGGNHIGGCDATSNLHKDGKLVPLL 121
> hsa:114112 TXNRD3, TGR, TR2, TRXR3; thioredoxin reductase 3
(EC:1.8.1.9); K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=607
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 54/100 (54%), Gaps = 5/100 (5%)
Query 58 QDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVS--DMHVEQIENNPHCNAIQD 115
+++ + + L+ +V IF+K++CP+ SL V + ++Q+++ +Q+
Sbjct 53 EELRRHLVGLIERSRVVIFSKSYCPHSTRVKELFSSLGVECNVLELDQVDDGAR---VQE 109
Query 116 YLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
L E T ++VP +F+N GG D T ++G LQKL+
Sbjct 110 VLSEITNQKTVPNIFVNKVHVGGCDQTFQAYQSGLLQKLL 149
> ath:AT5G63030 glutaredoxin, putative
Length=125
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 48/104 (46%), Gaps = 1/104 (0%)
Query 52 MALNSAQDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCN 111
M+ + V +V+ + V +F+KT+C YCQ L L + V +++
Sbjct 10 MSKEEMEVVVNKAKEIVSAYPVVVFSKTYCGYCQRVKQLLTQLGAT-FKVLELDEMSDGG 68
Query 112 AIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
IQ L E TG +VP VFI G GG D + K G L L+
Sbjct 69 EIQSALSEWTGQTTVPNVFIKGNHIGGCDRVMETNKQGKLVPLL 112
> ath:AT2G20270 glutaredoxin family protein
Length=179
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 48/93 (51%), Gaps = 0/93 (0%)
Query 64 VDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGA 123
V + V + V +++KT C Y + +SL V + VE + + +Q+ L++ TG
Sbjct 78 VKTTVAENPVVVYSKTWCSYSSQVKSLFKSLQVEPLVVELDQLGSEGSQLQNVLEKITGQ 137
Query 124 RSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIA 156
+VP VFI G GG DT+ G L+ ++A
Sbjct 138 YTVPNVFIGGKHIGGCSDTLQLHNKGELEAILA 170
> ath:AT3G54900 CXIP1; CXIP1 (CAX INTERACTING PROTEIN 1); antiporter/
glutathione disulfide oxidoreductase
Length=173
Score = 52.8 bits (125), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 51/104 (49%), Gaps = 12/104 (11%)
Query 60 VPQWVDSL---VNGHKVTIFAK-----THCPYCQAAIAALRSLNVSDMHVEQIENNPHCN 111
PQ D+L VN KV +F K C + + L++LNV V +EN
Sbjct 68 TPQLKDTLEKLVNSEKVVLFMKGTRDFPMCGFSNTVVQILKNLNVPFEDVNILENE---- 123
Query 112 AIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
++ LKE + + P+++I G FFGG D T+ K G LQ+ +
Sbjct 124 MLRQGLKEYSNWPTFPQLYIGGEFFGGCDITLEAFKTGELQEEV 167
> ath:AT3G62960 glutaredoxin family protein
Length=102
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 42/83 (50%), Gaps = 1/83 (1%)
Query 73 VTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFIN 132
V IF K+ C C A R L V + +I+N+P C I+ L A +VP VF++
Sbjct 13 VVIFTKSSCCLCYAVQILFRDLRVQPT-IHEIDNDPDCREIEKALVRLGCANAVPAVFVS 71
Query 133 GTFFGGGDDTVAGVKNGTLQKLI 155
G G +D ++ +G+L LI
Sbjct 72 GKLVGSTNDVMSLHLSGSLVPLI 94
> ath:AT2G47880 glutaredoxin family protein
Length=102
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 41/83 (49%), Gaps = 1/83 (1%)
Query 73 VTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFIN 132
V IF K+ C C A R L V + +I+N+P C I+ L + +VP VF+
Sbjct 13 VVIFTKSSCCLCYAVQILFRDLRVQPT-IHEIDNDPDCREIEKALLRLGCSTAVPAVFVG 71
Query 133 GTFFGGGDDTVAGVKNGTLQKLI 155
G G ++ ++ +G+L LI
Sbjct 72 GKLVGSTNEVMSLHLSGSLVPLI 94
> sce:YBR014C GRX7; Cis-golgi localized monothiol glutaredoxin;
more similar in activity to dithiol than other monothiol glutaredoxins;
involved in the oxidative stress response; does
not bind metal ions; functional overlap with GRX6
Length=203
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 42/79 (53%), Gaps = 2/79 (2%)
Query 75 IFAKTHCPYCQAAIAAL-RSLNVS-DMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFIN 132
+F+KT CPY + A L S S +V +++ + H +QD +++ TG R+VP V I
Sbjct 102 VFSKTGCPYSKKLKALLTNSYTFSPSYYVVELDRHEHTKELQDQIEKVTGRRTVPNVIIG 161
Query 133 GTFFGGGDDTVAGVKNGTL 151
GT GG + KN L
Sbjct 162 GTSRGGYTEIAELHKNDEL 180
> eco:b3610 grxC, ECK3600, JW3585, yibM; glutaredoxin 3; K03676
glutaredoxin 3
Length=83
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 29/83 (34%), Positives = 43/83 (51%), Gaps = 4/83 (4%)
Query 73 VTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFIN 132
V I+ K CPYC A A L S VS + I+ N A ++ + +++G +VP++FI+
Sbjct 4 VEIYTKETCPYCHRAKALLSSKGVSFQELP-IDGNA---AKREEMIKRSGRTTVPQIFID 59
Query 133 GTFFGGGDDTVAGVKNGTLQKLI 155
GG DD A G L L+
Sbjct 60 AQHIGGCDDLYALDARGGLDPLL 82
> hsa:7296 TXNRD1, GRIM-12, MGC9145, TR, TR1, TRXR1, TXNR; thioredoxin
reductase 1 (EC:1.8.1.9); K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=649
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/108 (27%), Positives = 48/108 (44%), Gaps = 5/108 (4%)
Query 48 FASTMALNSAQDVPQWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENN 107
F ST +S + ++D GH V IF+++ C C +SL V V +++
Sbjct 47 FTSTATADSRALLQAYID----GHSVVIFSRSTCTRCTEVKKLFKSLCVP-YFVLELDQT 101
Query 108 PHCNAIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
A++ L E +P VF+ GG T+ + G LQKL+
Sbjct 102 EDGRALEGTLSELAAETDLPVVFVKQRKIGGHGPTLKAYQEGRLQKLL 149
> hsa:51218 GLRX5, C14orf87, FLB4739, GRX5, MGC14129, PR01238,
PRO1238; glutaredoxin 5; K07390 monothiol glutaredoxin
Length=157
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 46/96 (47%), Gaps = 10/96 (10%)
Query 62 QWVDSLVNGHKVTIFAKT-----HCPYCQAAIAALRSLNVSDMHVEQIENNPHC-NAIQD 115
+ +D+LV KV +F K C + A + LR V D + ++P I+D
Sbjct 43 EQLDALVKKDKVVVFLKGTPEQPQCGFSNAVVQILRLHGVRDYAAYNVLDDPELRQGIKD 102
Query 116 YLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTL 151
Y + ++P+V++NG F GG D + +NG L
Sbjct 103 Y----SNWPTIPQVYLNGEFVGGCDILLQMHQNGDL 134
> mmu:73046 Glrx5, 2310004O13Rik, 2900070E19Rik, AU020725; glutaredoxin
5 homolog (S. cerevisiae); K07390 monothiol glutaredoxin
Length=152
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 45/94 (47%), Gaps = 10/94 (10%)
Query 64 VDSLVNGHKVTIFAKT-----HCPYCQAAIAALRSLNVSDMHVEQIENNPHC-NAIQDYL 117
+D+LV KV +F K C + A + LR V D + ++P I+DY
Sbjct 41 LDALVKKDKVVVFLKGTPEQPQCGFSNAVVQILRLHGVRDYAAYNVLDDPELRQGIKDY- 99
Query 118 KEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTL 151
+ ++P+V++NG F GG D + +NG L
Sbjct 100 ---SNWPTIPQVYLNGEFVGGCDILLQMHQNGDL 130
> dre:569013 MGC152951; zgc:152951
Length=372
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 26/76 (34%), Positives = 42/76 (55%), Gaps = 5/76 (6%)
Query 66 SLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARS 125
+L+ G +VT+++ CP+C A L +L V V ++ N H I+ +KE TG S
Sbjct 6 TLMKG-RVTVYSVPGCPHCTRAKTTLGALGVP---VCDVDVNKH-REIRALVKELTGHSS 60
Query 126 VPRVFINGTFFGGGDD 141
VP++F N + G +D
Sbjct 61 VPQIFFNNLYVGNNED 76
> xla:734675 glrx5, MGC116473, grx5; glutaredoxin 5; K07390 monothiol
glutaredoxin
Length=154
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 48/104 (46%), Gaps = 10/104 (9%)
Query 54 LNSAQDVPQWVDSLVNGHKVTIF-----AKTHCPYCQAAIAALRSLNVSDMHVEQI-ENN 107
L+S P+ +D LV KV +F A+ C + A + LR V+D + E+
Sbjct 32 LSSDSVSPEHLDGLVKKDKVVVFIKGTPAQPMCGFSNAVVQILRMHGVNDYAAYNVLEDQ 91
Query 108 PHCNAIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTL 151
I++Y T +P+V+ NG F GG D + +NG L
Sbjct 92 DLRQGIKNYSNWPT----IPQVYFNGEFVGGCDILLQMHQNGDL 131
> ath:AT2G38270 CXIP2; CXIP2 (CAX-INTERACTING PROTEIN 2); electron
carrier/ protein disulfide oxidoreductase
Length=293
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 45/99 (45%), Gaps = 6/99 (6%)
Query 62 QWVDSLVNGHKVTIFAK-----THCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDY 116
+ +D LV KV F K C + Q + L S V D + ++ + + +++
Sbjct 195 ELIDRLVKESKVVAFIKGSRSAPQCGFSQRVVGILESQGV-DYETVDVLDDEYNHGLRET 253
Query 117 LKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
LK + + P++F+ G GG D + +NG L ++
Sbjct 254 LKNYSNWPTFPQIFVKGELVGGCDILTSMYENGELANIL 292
> sce:YDL010W GRX6; Cis-golgi localized monothiol glutaredoxin
that binds an iron-sulfur cluster; more similar in activity
to dithiol than other monothiol glutaredoxins; involved in
the oxidative stress response; functional overlap with GRX7
Length=231
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/71 (25%), Positives = 39/71 (54%), Gaps = 2/71 (2%)
Query 73 VTIFAKTHCPYCQAAIAALRSLN--VSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVF 130
+ IF+K+ C Y + L + + + ++ +++ + H +Q+Y+K TG +VP +
Sbjct 128 IIIFSKSTCSYSKGMKELLENEYQFIPNYYIIELDKHGHGEELQEYIKLVTGRGTVPNLL 187
Query 131 INGTFFGGGDD 141
+NG GG ++
Sbjct 188 VNGVSRGGNEE 198
> ath:AT1G06830 glutaredoxin family protein
Length=99
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 39/83 (46%), Gaps = 1/83 (1%)
Query 73 VTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFIN 132
V IF K+ C A + L V+ + +I+ +P C I+ L ++ VP VFI
Sbjct 13 VVIFTKSSCCLSYAVQVLFQDLGVNP-KIHEIDKDPECREIEKALMRLGCSKPVPAVFIG 71
Query 133 GTFFGGGDDTVAGVKNGTLQKLI 155
G G ++ ++ + +L L+
Sbjct 72 GKLVGSTNEVMSMHLSSSLVPLV 94
> ath:AT2G30540 glutaredoxin family protein
Length=102
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 39/83 (46%), Gaps = 1/83 (1%)
Query 73 VTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFIN 132
V IF+K+ C A + L V V +I+ +P C I+ L + VP +F+
Sbjct 13 VVIFSKSSCCMSYAVQVLFQDLGVHPT-VHEIDKDPECREIEKALMRLGCSTPVPAIFVG 71
Query 133 GTFFGGGDDTVAGVKNGTLQKLI 155
G G ++ ++ +G+L L+
Sbjct 72 GKLIGSTNEVMSLHLSGSLVPLV 94
> ath:AT4G04950 thioredoxin family protein
Length=488
Score = 38.1 bits (87), Expect = 0.011, Method: Composition-based stats.
Identities = 25/119 (21%), Positives = 57/119 (47%), Gaps = 9/119 (7%)
Query 39 LRYNSCRTLFASTMALNSAQDVPQWVDSLVNGHKVTIFAK-----THCPYCQAAIAALRS 93
++ N+ +L +++A + ++ L N H V +F K C + + + L+
Sbjct 132 VKENAKASLQDRAQPVSTADALKSRLEKLTNSHPVMLFMKGIPEEPRCGFSRKVVDILKE 191
Query 94 LNVSDMHVEQIENNPHCNAIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQ 152
+NV + + +N +++ LK+ + + P+++ NG GG D +A ++G L+
Sbjct 192 VNVDFGSFDILSDN----EVREGLKKFSNWPTFPQLYCNGELLGGADIAIAMHESGELK 246
> pfa:PFC0205c PfGLP-1; 1-cys-glutaredoxin-like protein-1; K07390
monothiol glutaredoxin
Length=171
Score = 38.1 bits (87), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 25/99 (25%), Positives = 46/99 (46%), Gaps = 10/99 (10%)
Query 64 VDSLVNGHKVTIFAKTH-----CPYCQAAIAALRSLNVSD-MHVEQIENNPHCNAIQDYL 117
+ L+ K+ +F K C + + L S+NV D ++++ ++NN AI+ Y
Sbjct 77 IKELLEQEKIVLFMKGTPEKPLCGFSANVVNILNSMNVKDYVYIDVMKNNNLREAIKIY- 135
Query 118 KEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIA 156
+ +P +++N F GG D G L+K+I
Sbjct 136 ---SNWPYIPNLYVNNNFIGGYDIISDLYNRGELEKIIK 171
> bbo:BBOV_II007360 18.m06610; Thioredoxin-like protein 2
Length=202
Score = 37.4 bits (85), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 21/90 (23%), Positives = 47/90 (52%), Gaps = 7/90 (7%)
Query 55 NSAQDVPQWVDSLVNGHKVTIFAKTHC--PYCQAAIAALRSLNVSDMHVE--QIENNPHC 110
+S + + + L++ H++ +F K P+C+ + A + LN S + + I +P
Sbjct 102 DSGESTQERIKRLISTHRILLFMKGEKSDPFCRYSKAVVNMLNESGVEYDTYNIFEDPE- 160
Query 111 NAIQDYLKEKTGARSVPRVFINGTFFGGGD 140
+++ LK + + P++++NG+ GG D
Sbjct 161 --LREELKVYSNWPTYPQLYVNGSLIGGHD 188
> bbo:BBOV_II007430 18.m06616; thioredoxin-like protein 2
Length=202
Score = 37.4 bits (85), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 21/90 (23%), Positives = 47/90 (52%), Gaps = 7/90 (7%)
Query 55 NSAQDVPQWVDSLVNGHKVTIFAKTHC--PYCQAAIAALRSLNVSDMHVE--QIENNPHC 110
+S + + + L++ H++ +F K P+C+ + A + LN S + + I +P
Sbjct 102 DSGESTQERIKRLISTHRILLFMKGEKSDPFCRYSKAVVNMLNESGVEYDTYNIFEDPE- 160
Query 111 NAIQDYLKEKTGARSVPRVFINGTFFGGGD 140
+++ LK + + P++++NG+ GG D
Sbjct 161 --LREELKVYSNWPTYPQLYVNGSLIGGHD 188
> tpv:TP02_0196 hypothetical protein
Length=177
Score = 37.4 bits (85), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 27/110 (24%), Positives = 51/110 (46%), Gaps = 7/110 (6%)
Query 48 FASTMALNSAQDVPQWVDSLVNGHKVTIFAKTHC--PYCQAAIAALRSLNVSDMHVE--Q 103
F A + + + L+ H V +F K P+C+ + A + LN S + E
Sbjct 63 FVRGWAFKTRETTLDKIKRLLREHPVLLFMKGDKAEPFCRFSRAVVNMLNSSGVRFEGYN 122
Query 104 IENNPHCNAIQDYLKEKTGARSVPRVFINGTFFGGGDDTVAGVKNGTLQK 153
I ++P+ +++ LK + + P++++NG GG D + TL+K
Sbjct 123 IFDDPN---LREELKTYSNWPTYPQLYVNGNLIGGHDIIKELYETNTLRK 169
> ath:AT2G47870 glutaredoxin family protein
Length=103
Score = 37.4 bits (85), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 22/97 (22%), Positives = 45/97 (46%), Gaps = 2/97 (2%)
Query 62 QWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKT 121
+ V L + IF K+ C C + L S + +++ +P ++ L
Sbjct 2 ERVRDLASEKAAVIFTKSSCCMCHSIKTLFYELGASPA-IHELDKDPQGPDMERALFRVF 60
Query 122 GAR-SVPRVFINGTFFGGGDDTVAGVKNGTLQKLIAA 157
G+ +VP VF+ G + G D ++ +G+L++++ A
Sbjct 61 GSNPAVPAVFVGGRYVGSAKDVISFHVDGSLKQMLKA 97
> ath:AT5G06470 glutaredoxin family protein
Length=239
Score = 37.0 bits (84), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 122 GARSVPRVFINGTFFGGGDDTVAGVKNGTLQKLIAA 157
G + PR+FI G + GG ++ VA +NG L+KL+
Sbjct 143 GKVTSPRLFIRGRYIGGAEEVVALNENGKLKKLLQG 178
> ath:AT3G62950 glutaredoxin family protein
Length=103
Score = 35.8 bits (81), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 19/95 (20%), Positives = 44/95 (46%), Gaps = 2/95 (2%)
Query 62 QWVDSLVNGHKVTIFAKTHCPYCQAAIAALRSLNVSDMHVEQIENNPHCNAIQDYLKEKT 121
+ + L + IF K+ C C + L S + +++ +P ++ L+
Sbjct 2 ERIRDLSSKKAAVIFTKSSCCMCHSIKTLFYELGASPA-IHELDKDPEGREMERALRALG 60
Query 122 GAR-SVPRVFINGTFFGGGDDTVAGVKNGTLQKLI 155
+ +VP VF+ G + G D ++ +G+L++++
Sbjct 61 SSNPAVPAVFVGGRYIGSAKDIISFHVDGSLKQML 95
Lambda K H
0.324 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3516928200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40