bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2525_orf2
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
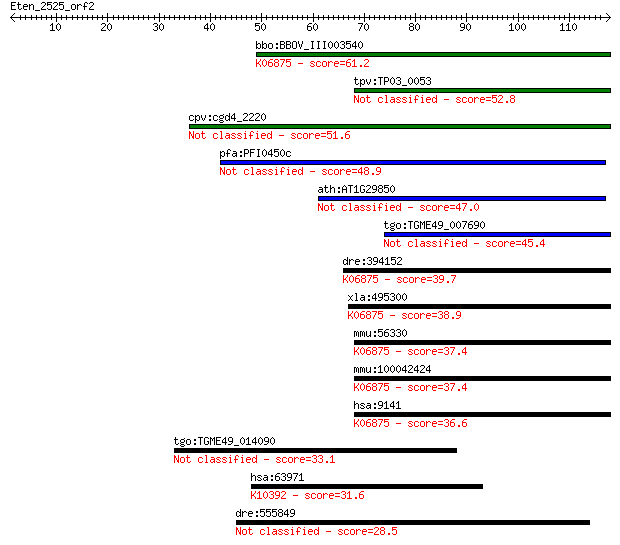
bbo:BBOV_III003540 17.m07333; hypothetical protein; K06875 pro... 61.2 8e-10
tpv:TP03_0053 hypothetical protein 52.8 3e-07
cpv:cgd4_2220 possible double-stranded DNA-binding domain, sma... 51.6 6e-07
pfa:PFI0450c PfARP; apoptosis-related protein, putative 48.9 4e-06
ath:AT1G29850 double-stranded DNA-binding family protein 47.0 2e-05
tgo:TGME49_007690 double-stranded DNA-binding domain-containin... 45.4 5e-05
dre:394152 pdcd5, MGC56315, zgc:56315; programmed cell death 5... 39.7 0.002
xla:495300 pdcd5; programmed cell death 5; K06875 programmed c... 38.9 0.004
mmu:56330 Pdcd5, 2200003D22Rik, AA408513, MGC103124, Tfar19; p... 37.4 0.013
mmu:100042424 Gm3837; predicted gene 3837; K06875 programmed c... 37.4 0.013
hsa:9141 PDCD5, FLJ42784, MGC9294, TFAR19; programmed cell dea... 36.6 0.023
tgo:TGME49_014090 hypothetical protein 33.1 0.22
hsa:63971 KIF13A, FLJ27232, bA500C11.2; kinesin family member ... 31.6 0.58
dre:555849 hmha1; histocompatibility (minor) HA-1 28.5 5.0
> bbo:BBOV_III003540 17.m07333; hypothetical protein; K06875 programmed
cell death protein 5
Length=125
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 48/74 (64%), Gaps = 5/74 (6%)
Query 49 MDAADANMQKQLQQQL-----QQQQQQEEKRKAIEEQRRMAMRSLLSPDAYQRLSRIHLV 103
MD D N Q Q + ++Q Q+E KR++I E RRM +R+LL+ +A +RL RI +V
Sbjct 1 MDGLDFNQAAQNAQAISHNTQEKQMQEEAKRESILEARRMTLRTLLTVEAQERLHRIAMV 60
Query 104 KPEKAMAAENYVMQ 117
KPEKA EN+++Q
Sbjct 61 KPEKATQIENFLLQ 74
> tpv:TP03_0053 hypothetical protein
Length=115
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/50 (48%), Positives = 38/50 (76%), Gaps = 0/50 (0%)
Query 68 QQQEEKRKAIEEQRRMAMRSLLSPDAYQRLSRIHLVKPEKAMAAENYVMQ 117
+Q+E KR+ + E RR+ +RS+L+ +A +RL+RI VKPEKA EN+++Q
Sbjct 19 EQEELKRQQLLEARRVGLRSILTVEAQERLNRISSVKPEKATLIENFLLQ 68
> cpv:cgd4_2220 possible double-stranded DNA-binding domain, small
conserved protein
Length=130
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 48/82 (58%), Gaps = 6/82 (7%)
Query 36 SSTSSTSSSSNGTMDAADANMQKQLQQQLQQQQQQEEKRKAIEEQRRMAMRSLLSPDAYQ 95
S + +S NG+ + ++ Q++ Q ++ IE+Q+R A+R++L + +
Sbjct 4 SDPNEFNSGKNGSNNVGPIETNQEFNQKVDDQ------KRIIEDQKRGALRAILENSSIE 57
Query 96 RLSRIHLVKPEKAMAAENYVMQ 117
RL+RI LVKP+K + E+Y+++
Sbjct 58 RLNRIALVKPDKVLQIEDYILR 79
> pfa:PFI0450c PfARP; apoptosis-related protein, putative
Length=131
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 52/75 (69%), Gaps = 4/75 (5%)
Query 42 SSSSNGTMDAADANMQKQLQQQLQQQQQQEEKRKAIEEQRRMAMRSLLSPDAYQRLSRIH 101
+ + + ++ + N++ + +++L+ +Q+QEE + E+RR+ ++SLL+P+A+ RLSRI
Sbjct 6 AEAKSKLIEMSKQNIENKTKKELELKQKQEE----LLEKRRLILKSLLTPEAHARLSRIA 61
Query 102 LVKPEKAMAAENYVM 116
+VK E+A E+ ++
Sbjct 62 IVKEEQARKIEDIII 76
> ath:AT1G29850 double-stranded DNA-binding family protein
Length=151
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 40/56 (71%), Gaps = 0/56 (0%)
Query 61 QQQLQQQQQQEEKRKAIEEQRRMAMRSLLSPDAYQRLSRIHLVKPEKAMAAENYVM 116
QQ +Q++QQE+ ++ +E+R+M + +LS A +R++RI LVKPEKA E+ ++
Sbjct 29 QQNPEQEKQQEDAKREADERRQMMLSQVLSSQARERIARIALVKPEKARGVEDVIL 84
> tgo:TGME49_007690 double-stranded DNA-binding domain-containing
protein
Length=242
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 21/44 (47%), Positives = 32/44 (72%), Gaps = 0/44 (0%)
Query 74 RKAIEEQRRMAMRSLLSPDAYQRLSRIHLVKPEKAMAAENYVMQ 117
R+ +EEQRR+ +R++L+P A +RL RI LVK +KA E ++Q
Sbjct 158 RQQMEEQRRIMLRAVLTPAAQERLHRIQLVKADKAREVEALILQ 201
> dre:394152 pdcd5, MGC56315, zgc:56315; programmed cell death
5; K06875 programmed cell death protein 5
Length=128
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/52 (46%), Positives = 32/52 (61%), Gaps = 1/52 (1%)
Query 66 QQQQQEEKRKAIEEQRRMAMRSLLSPDAYQRLSRIHLVKPEKAMAAENYVMQ 117
QQ QQE K++ E R + +L A RLS + LVKP+KA A ENY++Q
Sbjct 28 QQGQQEAKQRETE-MRNSILAQVLDQSARARLSNLALVKPDKAKAVENYLIQ 78
> xla:495300 pdcd5; programmed cell death 5; K06875 programmed
cell death protein 5
Length=126
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 23/51 (45%), Positives = 33/51 (64%), Gaps = 1/51 (1%)
Query 67 QQQQEEKRKAIEEQRRMAMRSLLSPDAYQRLSRIHLVKPEKAMAAENYVMQ 117
Q QQE K++ ++ R + +LS A RL+ + LVKPEKA A ENY++Q
Sbjct 28 QSQQEAKQRE-DDMRNNILAQVLSQAARARLNNLALVKPEKAKAVENYLIQ 77
> mmu:56330 Pdcd5, 2200003D22Rik, AA408513, MGC103124, Tfar19;
programmed cell death 5; K06875 programmed cell death protein
5
Length=126
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 22/50 (44%), Positives = 31/50 (62%), Gaps = 2/50 (4%)
Query 68 QQQEEKRKAIEEQRRMAMRSLLSPDAYQRLSRIHLVKPEKAMAAENYVMQ 117
QQ+ ++R+A E R + +L A RLS + LVKPEK A ENY++Q
Sbjct 29 QQEAKQREA--EMRNSILAQVLDQSARARLSNLALVKPEKTKAVENYLIQ 76
> mmu:100042424 Gm3837; predicted gene 3837; K06875 programmed
cell death protein 5
Length=126
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 22/50 (44%), Positives = 31/50 (62%), Gaps = 2/50 (4%)
Query 68 QQQEEKRKAIEEQRRMAMRSLLSPDAYQRLSRIHLVKPEKAMAAENYVMQ 117
QQ+ ++R+A E R + +L A RLS + LVKPEK A ENY++Q
Sbjct 29 QQEAKQREA--EMRNSILAQVLDQSARARLSNLALVKPEKTKAVENYLIQ 76
> hsa:9141 PDCD5, FLJ42784, MGC9294, TFAR19; programmed cell death
5; K06875 programmed cell death protein 5
Length=125
Score = 36.6 bits (83), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 22/50 (44%), Positives = 30/50 (60%), Gaps = 2/50 (4%)
Query 68 QQQEEKRKAIEEQRRMAMRSLLSPDAYQRLSRIHLVKPEKAMAAENYVMQ 117
QQ+ + R+A E R + +L A RLS + LVKPEK A ENY++Q
Sbjct 29 QQEAKHREA--EMRNSILAQVLDQSARARLSNLALVKPEKTKAVENYLIQ 76
> tgo:TGME49_014090 hypothetical protein
Length=1859
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 33/55 (60%), Gaps = 2/55 (3%)
Query 33 RSSSSTSSTSSSSNGTMDAADANMQKQLQQQLQQQQQQEEKRKAIEEQRRMAMRS 87
R +S +T +S +G D A ++KQ ++Q Q Q+Q + + +EEQ RMA RS
Sbjct 929 RRASGAEATGASDDG--DPVLAQLEKQEERQRQLQEQLNQVKSQLEEQLRMATRS 981
> hsa:63971 KIF13A, FLJ27232, bA500C11.2; kinesin family member
13A; K10392 kinesin family member 1/13/14
Length=1770
Score = 31.6 bits (70), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 27/51 (52%), Gaps = 6/51 (11%)
Query 48 TMDAADANMQKQLQQQLQQQQQQEEKRKAIEEQRRMAMRSL------LSPD 92
M ++N Q Q+ ++Q EEKR A+EEQR M R L LSPD
Sbjct 588 IMKTLNSNDPVQNVVQVLEKQYLEEKRSALEEQRLMYERELEQLRQQLSPD 638
> dre:555849 hmha1; histocompatibility (minor) HA-1
Length=1127
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 41/75 (54%), Gaps = 7/75 (9%)
Query 45 SNGTMDAADANMQKQ--LQQQLQQQQQQEEKRKAIEEQRRMAMRSLLSPDA-YQRLSRIH 101
SNG M A A +Q Q + + ++Q+ E+KRK I+EQ A R L+ D +R +++
Sbjct 373 SNG-MHQAAATLQNQTFIMPLMVRKQEHEKKRKEIKEQWLKAKRKLMECDVNLRRAKQVY 431
Query 102 LVKPE---KAMAAEN 113
+ + E KA A N
Sbjct 432 MTRCEEYDKARTAAN 446
Lambda K H
0.306 0.115 0.304
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2032807080
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40