bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2531_orf1
Length=195
Score E
Sequences producing significant alignments: (Bits) Value
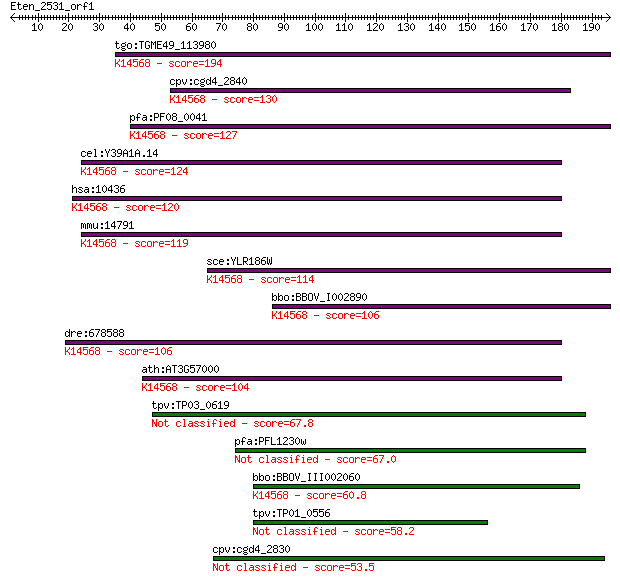
tgo:TGME49_113980 nucleolar essential protein 1, putative ; K1... 194 1e-49
cpv:cgd4_2840 Mra1/NEP1 like protein, involved in pre-rRNA pro... 130 3e-30
pfa:PF08_0041 ribosome biogenesis protein nep1 homologue, puta... 127 2e-29
cel:Y39A1A.14 hypothetical protein; K14568 essential for mitot... 124 2e-28
hsa:10436 EMG1, C2F, FLJ60792, Grcc2f, NEP1; EMG1 nucleolar pr... 120 4e-27
mmu:14791 Emg1, C2f, Grcc2f; EMG1 nucleolar protein homolog (S... 119 6e-27
sce:YLR186W EMG1, NEP1; Emg1p; K14568 essential for mitotic gr... 114 2e-25
bbo:BBOV_I002890 19.m02290; suppressor Mra1 family domain cont... 106 5e-23
dre:678588 MGC136360; zgc:136360; K14568 essential for mitotic... 106 6e-23
ath:AT3G57000 nucleolar essential protein-related; K14568 esse... 104 2e-22
tpv:TP03_0619 hypothetical protein 67.8 2e-11
pfa:PFL1230w conserved Plasmodium protein 67.0 4e-11
bbo:BBOV_III002060 17.m07199; hypothetical protein; K14568 ess... 60.8 2e-09
tpv:TP01_0556 hypothetical protein 58.2 2e-08
cpv:cgd4_2830 Mra1/NEP1 like protein, involved in pre-rRNA pro... 53.5 4e-07
> tgo:TGME49_113980 nucleolar essential protein 1, putative ;
K14568 essential for mitotic growth 1
Length=235
Score = 194 bits (494), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 95/161 (59%), Positives = 123/161 (76%), Gaps = 0/161 (0%)
Query 35 RADRLQGRKVVVLLEDAVLDLAKGREGSLQLLEGFQHRKLLQQENRSIADVRPDITHQCL 94
R RL G++V+V+LE A L+LA+G++ SLQLL +H++LL++ +R +VRPDI H CL
Sbjct 19 RERRLTGQRVIVVLEGASLELAQGKDRSLQLLNSLEHKQLLRKCDRQGDEVRPDIAHHCL 78
Query 95 IALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKN 154
++LQESPLN+AGRLCVFIRT QLIE+ P L VPPTY EF KLM NLL+ RR++AVEKN
Sbjct 79 LSLQESPLNRAGRLCVFIRTADRQLIEISPLLTVPPTYQEFAKLMTNLLYARRLKAVEKN 138
Query 155 VTLMQVTKNDANLFLPVGSRKIALSTQGRPVDLGDFVQQFQ 195
VTL Q+ KN+ FLP S K+AL+ GR V L DFVQ+F+
Sbjct 139 VTLAQIVKNEHANFLPPNSVKVALTVSGRSVALSDFVQRFK 179
> cpv:cgd4_2840 Mra1/NEP1 like protein, involved in pre-rRNA processing,
adjacent genes paralogs ; K14568 essential for mitotic
growth 1
Length=223
Score = 130 bits (327), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 68/142 (47%), Positives = 91/142 (64%), Gaps = 12/142 (8%)
Query 53 LDLAKGREGSLQLLEGFQHRKLLQQENRS------------IADVRPDITHQCLIALQES 100
L+L + ++ SL+LL G H K +E S I ++RPDI HQCL++L +S
Sbjct 7 LELYEKKDKSLELLNGIDHPKKKIEEYCSLYSSKIFSKEEMILNIRPDILHQCLLSLLDS 66
Query 101 PLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQV 160
PLNK+GRL V+IRT S LIEV+PQL VP ++ EF LM+NLL R+I AV N +LM++
Sbjct 67 PLNKSGRLLVYIRTMSNVLIEVNPQLSVPRSFKEFSSLMVNLLVKRKITAVNGNTSLMRI 126
Query 161 TKNDANLFLPVGSRKIALSTQG 182
KND + LPVG +K LS G
Sbjct 127 VKNDIDKILPVGGKKYGLSLNG 148
> pfa:PF08_0041 ribosome biogenesis protein nep1 homologue, putative;
K14568 essential for mitotic growth 1
Length=279
Score = 127 bits (319), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 59/159 (37%), Positives = 99/159 (62%), Gaps = 3/159 (1%)
Query 40 QGRKVVVLLEDAVLDLAKGREGSLQLLEGFQHRKLLQQ---ENRSIADVRPDITHQCLIA 96
+ +KV+++LE A L L + + +L +H+ +L++ + +I + RPDI HQCLI
Sbjct 65 KKKKVIIILEGACLQLIEVKRFIYELANSRKHKNILKKNKVDEENIKNFRPDILHQCLIH 124
Query 97 LQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVT 156
L ESPLNK G L ++I+T QL V L++P T+ +F LM+ L +I+A EKN+
Sbjct 125 LLESPLNKFGYLQIYIKTHDNQLFYVSSNLKIPKTFQQFESLMVTFLRKYKIKANEKNIY 184
Query 157 LMQVTKNDANLFLPVGSRKIALSTQGRPVDLGDFVQQFQ 195
L+++ KND LP+ KI LS +G+ V+L ++++ ++
Sbjct 185 LLKIIKNDLQNILPINGHKIGLSLKGKKVELNNYIKVYK 223
> cel:Y39A1A.14 hypothetical protein; K14568 essential for mitotic
growth 1
Length=231
Score = 124 bits (310), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 69/159 (43%), Positives = 98/159 (61%), Gaps = 4/159 (2%)
Query 24 WEQVAP--RAEVKRADRLQGRKVV-VLLEDAVLDLAKGREGSLQLLEGFQHRKLLQQENR 80
++ VAP +K ++L+ +K++ V+LE L+ AK G +L +H L+++ +
Sbjct 5 YDTVAPPNAKRMKTDNQLEDKKILYVVLEGCSLETAK-VGGEYAILSSDKHANFLRKQKK 63
Query 81 SIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMI 140
AD RPDI HQCL+ L +SPLN+AG+L VF RT L++V PQ R+P T+D F LM+
Sbjct 64 DPADYRPDILHQCLLNLLDSPLNRAGKLRVFFRTSKNVLVDVSPQCRIPRTFDRFCGLMV 123
Query 141 NLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALS 179
LLH IRA E LM V KN + LPVGSRK+ +S
Sbjct 124 QLLHKLSIRAAETTQKLMSVVKNPVSNHLPVGSRKMLMS 162
> hsa:10436 EMG1, C2F, FLJ60792, Grcc2f, NEP1; EMG1 nucleolar
protein homolog (S. cerevisiae); K14568 essential for mitotic
growth 1
Length=244
Score = 120 bits (300), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 64/160 (40%), Positives = 98/160 (61%), Gaps = 2/160 (1%)
Query 21 AEMWEQVAP-RAEVKRADRLQGRKVVVLLEDAVLDLAKGREGSLQLLEGFQHRKLLQQEN 79
A+ W+ + P R + +++ GR+++V+LE A L+ K + + +LL +H+ +L +
Sbjct 19 AQDWDALPPKRPRLGAGNKIGGRRLIVVLEGASLETVKVGK-TYELLNCDKHKSILLKNG 77
Query 80 RSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLM 139
R + RPDITHQ L+ L +SPLN+AG L V+I TQ LIEV+PQ R+P T+D F LM
Sbjct 78 RDPGEARPDITHQSLLMLMDSPLNRAGLLQVYIHTQKNVLIEVNPQTRIPRTFDRFCGLM 137
Query 140 INLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALS 179
+ LLH +RA + L++V KN + PVG K+ S
Sbjct 138 VQLLHKLSVRAADGPQKLLKVIKNPVSDHFPVGCMKVGTS 177
> mmu:14791 Emg1, C2f, Grcc2f; EMG1 nucleolar protein homolog
(S. cerevisiae); K14568 essential for mitotic growth 1
Length=244
Score = 119 bits (298), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 66/158 (41%), Positives = 95/158 (60%), Gaps = 4/158 (2%)
Query 24 WEQVAPRA-EVKRADRLQGRKVVVLLEDAVLDLAK-GREGSLQLLEGFQHRKLLQQENRS 81
WE P+ + + GR+++V+LE A L+ K G+ + +LL +H+ +L + R
Sbjct 22 WETTPPKKLRLGAGSKCGGRRLIVVLEGASLETVKVGK--TYELLNCDRHKSMLLKNGRD 79
Query 82 IADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMIN 141
+VRPDITHQ L+ L +SPLN+AG L V+I TQ LIEV+PQ R+P T+D F LM+
Sbjct 80 PGEVRPDITHQSLLMLMDSPLNRAGLLQVYIHTQKNVLIEVNPQTRIPRTFDRFCGLMVQ 139
Query 142 LLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALS 179
LLH +RA + L++V KN + PVG KI S
Sbjct 140 LLHKLSVRAADGPQKLLKVIKNPVSDHFPVGCMKIGTS 177
> sce:YLR186W EMG1, NEP1; Emg1p; K14568 essential for mitotic
growth 1
Length=252
Score = 114 bits (285), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 56/131 (42%), Positives = 82/131 (62%), Gaps = 0/131 (0%)
Query 65 LLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHP 124
LL H+ LL++ R I++ RPDITHQCL+ L +SP+NKAG+L V+I+T G LIEV+P
Sbjct 67 LLNCDDHQGLLKKMGRDISEARPDITHQCLLTLLDSPINKAGKLQVYIQTSRGILIEVNP 126
Query 125 QLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALSTQGRP 184
+R+P T+ F LM+ LLH IR+V L++V KN LP RK+ LS
Sbjct 127 TVRIPRTFKRFSGLMVQLLHKLSIRSVNSEEKLLKVIKNPITDHLPTKCRKVTLSFDAPV 186
Query 185 VDLGDFVQQFQ 195
+ + D++++
Sbjct 187 IRVQDYIEKLD 197
> bbo:BBOV_I002890 19.m02290; suppressor Mra1 family domain containing
protein; K14568 essential for mitotic growth 1
Length=188
Score = 106 bits (264), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 53/110 (48%), Positives = 70/110 (63%), Gaps = 0/110 (0%)
Query 86 RPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMINLLHT 145
R DI HQ L+ L +SPLNK G L V IRT G L+EV P LRVP T +F L++ LL+
Sbjct 23 RVDILHQSLLVLLDSPLNKNGFLKVLIRTDDGNLVEVSPHLRVPRTPKQFESLLVYLLYK 82
Query 146 RRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALSTQGRPVDLGDFVQQFQ 195
R +++ E++ LM+ KN+ + LP GSR+ LS GR V L DF QF+
Sbjct 83 RHVKSQERDAILMRFIKNEIEIALPPGSRRFGLSVGGRQVKLKDFCHQFK 132
> dre:678588 MGC136360; zgc:136360; K14568 essential for mitotic
growth 1
Length=238
Score = 106 bits (264), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 61/165 (36%), Positives = 100/165 (60%), Gaps = 6/165 (3%)
Query 19 RSAEMWEQVAPR-AEVKRA--DRLQGRKVVVLLEDAVLDLAK-GREGSLQLLEGFQHRKL 74
R E ++ P+ A+ +R+ D++ +++VV+LE A L+ K G+ + +LL QH+ +
Sbjct 9 RGLEHLDEYEPKQAKQQRSLHDQMTEKRLVVVLEGATLETVKVGK--TFELLNCDQHKSM 66
Query 75 LQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDE 134
+ + R +RPDI HQCL+ L +SPLN+AG L V+I T+ LIE++PQ R+P T+
Sbjct 67 IIKSGRDPGKIRPDIAHQCLLMLLDSPLNRAGLLQVYIHTEKNVLIEINPQTRIPRTFAR 126
Query 135 FRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALS 179
F LM+ LLH +RA + L+++ KN + LP G + + S
Sbjct 127 FCGLMVQLLHKLSVRAADGPQRLLRLIKNPVSDHLPPGCPRFSTS 171
> ath:AT3G57000 nucleolar essential protein-related; K14568 essential
for mitotic growth 1
Length=298
Score = 104 bits (259), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 60/139 (43%), Positives = 84/139 (60%), Gaps = 5/139 (3%)
Query 44 VVVLLEDAVLDLAK-GREGSLQLLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPL 102
+V +LE A L++AK G+ + QLL H L++ NR+ AD RPDITHQ L+ + +SP+
Sbjct 91 IVFVLEKASLEVAKVGK--TYQLLNSDDHANFLKKNNRNPADYRPDITHQALLMILDSPV 148
Query 103 NKAGRL-CVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVT 161
NKAGRL V++RT+ G L EV P +R+P T+ F +M+ LL I AV L++
Sbjct 149 NKAGRLKAVYVRTEKGVLFEVKPHVRIPRTFKRFAGIMLQLLQKLSITAVNSREKLLRCV 208
Query 162 KND-ANLFLPVGSRKIALS 179
KN LPV S +I S
Sbjct 209 KNPIEEHHLPVNSHRIGFS 227
> tpv:TP03_0619 hypothetical protein
Length=271
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 43/147 (29%), Positives = 78/147 (53%), Gaps = 9/147 (6%)
Query 47 LLEDAVLDLAKGREGS----LQLLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPL 102
++ ++ L+ K + S L LL+ H ++ E+R+ +D RPD+ H CL++LQ+S L
Sbjct 6 VISNSGLEFHKSNKKSPHPKLNLLQEHLHFNNIE-EDRNGSDYRPDLVHFCLLSLQDSIL 64
Query 103 NKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMINLLHTRR--IRAVEKNVTLMQV 160
NK G++ V++ T G + +V RVP + F K+ LH++ +R ++ + V
Sbjct 65 NKEGKIQVYLETLQGHVFKVSNSFRVPRAFKVFNKVFSTFLHSKSKDLRTESGDILIESV 124
Query 161 TKNDANLFLPVGSRKIALSTQGRPVDL 187
D +P S KIA+ + +D+
Sbjct 125 VNLDQ--LIPDSSIKIAIDNRCPVLDI 149
> pfa:PFL1230w conserved Plasmodium protein
Length=443
Score = 67.0 bits (162), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 39/114 (34%), Positives = 67/114 (58%), Gaps = 2/114 (1%)
Query 74 LLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYD 133
++Q + ++R DI L++L++S LNK G+L ++I T +G I V P RVP +
Sbjct 264 IIQSVKNKLENIRLDILFFTLLSLRDSILNKKGKLQIYIYTVNGSFIFVSPLFRVPRNFT 323
Query 134 EFRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALSTQGRPVDL 187
F+K+M+NLL + +KNV L+++ ND ++ + IA+S +G P D+
Sbjct 324 LFKKVMLNLLKRGVVYDDQKNV-LLKIIFNDITSYVE-DAVCIAISNKGFPTDV 375
> bbo:BBOV_III002060 17.m07199; hypothetical protein; K14568 essential
for mitotic growth 1
Length=293
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 36/116 (31%), Positives = 62/116 (53%), Gaps = 18/116 (15%)
Query 80 RSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLM 139
R+ +VRPDI H CL+A+ +S LNK + +++ T + Q+ + R+P T+ F K+M
Sbjct 74 RNWKEVRPDILHFCLLAIHDSILNKEKLVQIWVHTLNSQIYRFSSEFRIPRTFKVFNKVM 133
Query 140 INLLHTRRIRAVEKNVTLMQVTKNDANLF-----LPVGSR-----KIALSTQGRPV 185
LH+ N ++ +D+ +F LP+ + KIA+S +GRP+
Sbjct 134 AKYLHS--------NAKCLKPEGDDSTVFVERMDLPLDNLGNNGIKIAVSNRGRPL 181
> tpv:TP01_0556 hypothetical protein
Length=105
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/76 (39%), Positives = 47/76 (61%), Gaps = 0/76 (0%)
Query 80 RSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLM 139
R+ + R DI HQ L+ LQ+S LNK L V+IRT +LI+V+P VP T + F L+
Sbjct 13 RNTRESRLDILHQSLLILQDSILNKELCLKVYIRTVDNELIDVNPAFSVPRTLELFEVLI 72
Query 140 INLLHTRRIRAVEKNV 155
+L+ R++++ N+
Sbjct 73 QSLVTNRKVKSTNNNI 88
> cpv:cgd4_2830 Mra1/NEP1 like protein, involved in pre-rRNA processing,
adjacent genes paralogs
Length=216
Score = 53.5 bits (127), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 38/133 (28%), Positives = 63/133 (47%), Gaps = 6/133 (4%)
Query 67 EGFQHRKLLQQENRSIA--DVRPDITHQCLIALQESPLNKAGRLC-VFIRTQSGQLIEVH 123
E K+L+++ + I + RPDI H L+ L +SPL K+G L + I G+LI V
Sbjct 20 ELLTEEKVLERDMKLIDSDEFRPDIIHNSLLMLLDSPLCKSGCLSDILILNLDGKLIRVS 79
Query 124 PQLRVPPTYDEFRKLMINLLHTRRIRAV---EKNVTLMQVTKNDANLFLPVGSRKIALST 180
P+ RVP ++ F K+ L + E+N L+ + + +L + LS
Sbjct 80 PKFRVPRSFKVFSKVFSEFLSSPNGELKLPDEENTVLISLLNSSIEEYLSNSEVVVGLSR 139
Query 181 QGRPVDLGDFVQQ 193
+ V L +F +
Sbjct 140 MAKKVSLQEFCRN 152
Lambda K H
0.322 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5673387808
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40