bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2533_orf1
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
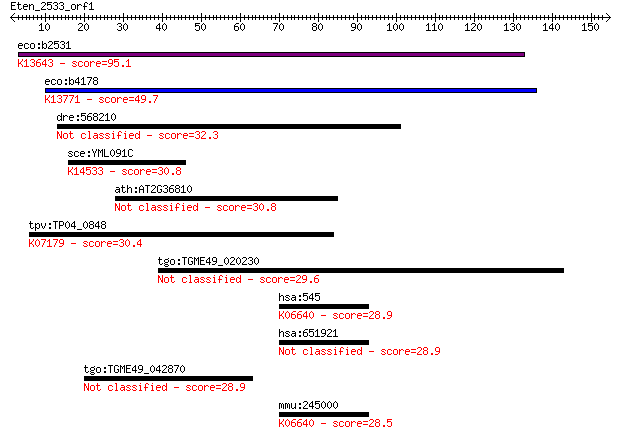
eco:b2531 iscR, ECK2528, JW2515, yfhP; DNA-binding transcripti... 95.1 7e-20
eco:b4178 nsrR, ECK4174, JW4136, yjeB; nitric oxide-sensitive ... 49.7 3e-06
dre:568210 Ral GTPase-activating protein alpha subunit 2-like 32.3 0.61
sce:YML091C RPM2; Protein subunit of mitochondrial RNase P, ha... 30.8 1.6
ath:AT2G36810 binding 30.8 1.8
tpv:TP04_0848 hypothetical protein; K07179 RIO kinase 2 [EC:2.... 30.4 2.6
tgo:TGME49_020230 leucine rich repeat protein, putative (EC:2.... 29.6 4.5
hsa:545 ATR, FRP1, MEC1, SCKL, SCKL1; ataxia telangiectasia an... 28.9 6.4
hsa:651921 serine/threonine-protein kinase ATR-like 28.9 7.0
tgo:TGME49_042870 hypothetical protein 28.9 7.0
mmu:245000 Atr; ataxia telangiectasia and Rad3 related (EC:2.7... 28.5 8.7
> eco:b2531 iscR, ECK2528, JW2515, yfhP; DNA-binding transcriptional
repressor; K13643 Rrf2 family transcriptional regulator,
iron-sulfur cluster assembly transcription factor
Length=162
Score = 95.1 bits (235), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 51/130 (39%), Positives = 76/130 (58%), Gaps = 2/130 (1%)
Query 3 KVSTKGRYALRTMIDLTVNNTGNPIPLKDIATRQNISLKYMEQIISLLIKAKLVKSVRGN 62
++++KGRYA+ M+D+ +N+ P+PL DI+ RQ ISL Y+EQ+ S L K LV SVRG
Sbjct 2 RLTSKGRYAVTAMLDVALNSEAGPVPLADISERQGISLSYLEQLFSRLRKNGLVSSVRGP 61
Query 63 NGGYLLAKSSMQYTAGDILRAAEGDMAPIACIEKDHCNCNRADYCSVLPFWLGLNKVING 122
GGYLL K + G+++ A D + A + C D C W L+ + G
Sbjct 62 GGGYLLGKDASSIAVGEVISAV--DESVDATRCQGKGGCQGGDKCLTHALWRDLSDRLTG 119
Query 123 YLDSVTLSEL 132
+L+++TL EL
Sbjct 120 FLNNITLGEL 129
> eco:b4178 nsrR, ECK4174, JW4136, yjeB; nitric oxide-sensitive
repressor for NO regulon; K13771 Rrf2 family transcriptional
regulator, nitric oxide-sensitive transcriptional repressor
Length=141
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 35/132 (26%), Positives = 66/132 (50%), Gaps = 16/132 (12%)
Query 10 YALRTMIDLTVNNTGNPIPLKDIATRQNISLKYMEQIISLLIKAKLVKSVRGNNGGYLLA 69
Y LR +I + G + ++ +S +M +II+ L +A V +VRG NGG L
Sbjct 9 YGLRALIYMASLPEGRMTSISEVTDVYGVSRNHMVKIINQLSRAGYVTAVRGKNGGIRLG 68
Query 70 KSSMQYTAGDILRAAEGDMAPIACIEKDHCNCNRADYCSVLP---FWLGLNKVINGY--- 123
K + GD++R ++ P++ + NC+ +++C + P L+K + +
Sbjct 69 KPASAIRIGDVVR----ELEPLSLV-----NCS-SEFCHITPACRLKQALSKAVQSFLTE 118
Query 124 LDSVTLSELARQ 135
LD+ TL++L +
Sbjct 119 LDNYTLADLVEE 130
> dre:568210 Ral GTPase-activating protein alpha subunit 2-like
Length=2050
Score = 32.3 bits (72), Expect = 0.61, Method: Composition-based stats.
Identities = 28/104 (26%), Positives = 45/104 (43%), Gaps = 19/104 (18%)
Query 13 RTMIDLTVNNTGNPIPLKDIATRQNIS------------LKYMEQIISLLIKAKLVKSVR 60
+ +DL ++ P PL TR N S + ++ +++ ++ K V S
Sbjct 265 KPQLDLPIHR---PKPLYVPVTRNNESTYCTRDQYLAPRVAFITWLVNFFLEKKYVNSTT 321
Query 61 GN--NGGYLLAKSSMQYTAGDILRAAEGDMAPIACI--EKDHCN 100
N NGG +L K TAG I + D+ P + EK+H N
Sbjct 322 SNSKNGGEVLPKLIQTVTAGSISQEKSTDLEPNGPVEQEKNHSN 365
> sce:YML091C RPM2; Protein subunit of mitochondrial RNase P,
has roles in nuclear transcription, cytoplasmic and mitochondrial
RNA processing, and mitochondrial translation; distributed
to mitochondria, cytoplasmic processing bodies, and the
nucleus (EC:3.1.26.5); K14533 ribonuclease P protein component,
mitochondrial [EC:3.1.26.5]
Length=1202
Score = 30.8 bits (68), Expect = 1.6, Method: Composition-based stats.
Identities = 11/30 (36%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 16 IDLTVNNTGNPIPLKDIATRQNISLKYMEQ 45
+D ++NNT NP+ +++ QN+SL+ ++Q
Sbjct 75 LDSSINNTNNPLTHEELLYNQNVSLRSLKQ 104
> ath:AT2G36810 binding
Length=1716
Score = 30.8 bits (68), Expect = 1.8, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 35/61 (57%), Gaps = 6/61 (9%)
Query 28 PLKD----IATRQNISLKYMEQIISLLIKAKLVKSVRGNNGGYLLAKSSMQYTAGDILRA 83
P++D + +S+ +ME +IS+L ++ LVKS ++ G + SS + DIL+A
Sbjct 1213 PMQDAFYAFSQHTELSVLFMEHLISILNRSSLVKS--DSHKGENTSSSSETHVEDDILQA 1270
Query 84 A 84
A
Sbjct 1271 A 1271
> tpv:TP04_0848 hypothetical protein; K07179 RIO kinase 2 [EC:2.7.11.1]
Length=467
Score = 30.4 bits (67), Expect = 2.6, Method: Composition-based stats.
Identities = 26/78 (33%), Positives = 37/78 (47%), Gaps = 4/78 (5%)
Query 6 TKGRYALRTMIDLTVNNTGNPIPLKDIATRQNISLKYMEQIISLLIKAKLVKSVRGNNGG 65
T + + T I++ + N IPL I T N+ M ++IS L+KAKL+ G
Sbjct 12 TNTEFRVLTAIEIGMRNH-QYIPLSLITTTANLRSVGMSKVISSLLKAKLIVHCGKVYDG 70
Query 66 YLLAKSSMQYTAGDILRA 83
Y L + Y A LRA
Sbjct 71 YKLTFLGLDYLA---LRA 85
> tgo:TGME49_020230 leucine rich repeat protein, putative (EC:2.7.11.1
3.1.3.16)
Length=1289
Score = 29.6 bits (65), Expect = 4.5, Method: Composition-based stats.
Identities = 31/112 (27%), Positives = 51/112 (45%), Gaps = 15/112 (13%)
Query 39 SLKYMEQIISLLIKAKLVKSVRGNNGGYLLAKSSMQYTAGDILRAAEGDMAPIACIEKDH 98
S+ Y +Q++ +L +A ++G + LA + TA + R EG +P +E+ +
Sbjct 542 SVHYADQLVDILAEA-----LKGTSSLRSLACAGNGMTAVGLRRLCEGLASPTCRLEELN 596
Query 99 CNCNRADYCSVLPF--WLGLNKVI------NGYLDSVTLSELARQAKENNAM 142
CN D VLP L N+ I N L S + +LA +EN +
Sbjct 597 VACN--DLQEVLPLAELLLTNRTIRILDVANCMLPSEAVDQLAAALEENTTL 646
> hsa:545 ATR, FRP1, MEC1, SCKL, SCKL1; ataxia telangiectasia
and Rad3 related (EC:2.7.11.1); K06640 ataxia telangiectasia
and Rad3 related [EC:2.7.11.1]
Length=2644
Score = 28.9 bits (63), Expect = 6.4, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 70 KSSMQYTAGDILRAAEGDMAPIA 92
K ++ T GDI RAA+GD+ P A
Sbjct 866 KDTLILTTGDIGRAAKGDLVPFA 888
> hsa:651921 serine/threonine-protein kinase ATR-like
Length=2105
Score = 28.9 bits (63), Expect = 7.0, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 70 KSSMQYTAGDILRAAEGDMAPIA 92
K ++ T GDI RAA+GD+ P A
Sbjct 624 KDTLILTTGDIGRAAKGDLVPFA 646
> tgo:TGME49_042870 hypothetical protein
Length=361
Score = 28.9 bits (63), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 20 VNNTGNPIPLKDIATRQNISLKYMEQIISLLIKAKLVKSVRGN 62
+ N L+DIA ++ I L Y+++ + L + +L+KS+ N
Sbjct 289 LTNAAQVTALRDIAEKEEILLSYIDESLPLAERQRLLKSMSQN 331
> mmu:245000 Atr; ataxia telangiectasia and Rad3 related (EC:2.7.11.1);
K06640 ataxia telangiectasia and Rad3 related [EC:2.7.11.1]
Length=2641
Score = 28.5 bits (62), Expect = 8.7, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 70 KSSMQYTAGDILRAAEGDMAPIA 92
K ++ T GDI RAA+GD+ P A
Sbjct 866 KDTLILTTGDIGRAAKGDLIPFA 888
Lambda K H
0.316 0.131 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3321543300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40