bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
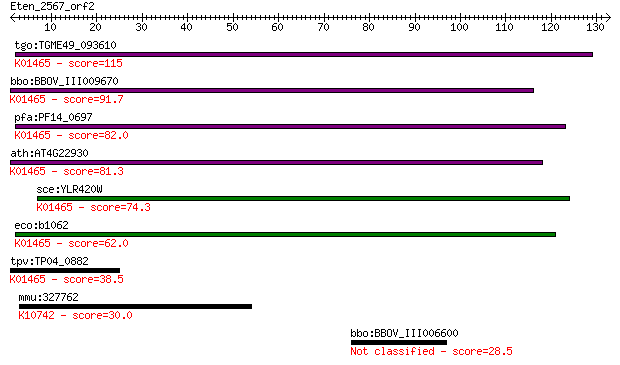
Query= Eten_2567_orf2
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_093610 dihydroorotase protein, putative (EC:3.5.2.3... 115 3e-26
bbo:BBOV_III009670 17.m07839; dihydroorotase (EC:3.5.2.3); K01... 91.7 6e-19
pfa:PF14_0697 dihydroorotase, putative; K01465 dihydroorotase ... 82.0 4e-16
ath:AT4G22930 PYR4; PYR4 (PYRIMIDIN 4); dihydroorotase/ hydrol... 81.3 8e-16
sce:YLR420W URA4; Dihydroorotase, catalyzes the third enzymati... 74.3 8e-14
eco:b1062 pyrC, ECK1047, JW1049; dihydro-orotase (EC:3.5.2.3);... 62.0 5e-10
tpv:TP04_0882 dihydroorotase (EC:3.5.2.3); K01465 dihydroorota... 38.5 0.006
mmu:327762 Dna2, Dna2l, E130315B21Rik, KIAA0083; DNA replicati... 30.0 2.1
bbo:BBOV_III006600 17.m07584; hypothetical protein 28.5 6.2
> tgo:TGME49_093610 dihydroorotase protein, putative (EC:3.5.2.3);
K01465 dihydroorotase [EC:3.5.2.3]
Length=403
Score = 115 bits (289), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 68/127 (53%), Positives = 85/127 (66%), Gaps = 0/127 (0%)
Query 2 QVVREGHPQFFLGSDSAPHPRHKKDSYPPAAGVFTQPFLLAYLADTFARAGCLDKLQSFA 61
QV+++G FFLGSDSAPHPR K+S PPAAGVFTQP LLAYL FA GC+ KL+ FA
Sbjct 270 QVIQDGDTHFFLGSDSAPHPRLAKESSPPAAGVFTQPLLLAYLVSIFAELGCISKLREFA 329
Query 62 CENAAAFFGLPKKELVEGEDCVVLEQTPCRIPNVVCGAEGTSTDVVPFLAGHELGYTLKV 121
+AAAFFG L E +C V++ TP R+P++ + VVPF+A EL YT++V
Sbjct 330 DGHAAAFFGFEAATLEENLECAVIQPTPQRVPSIFKFPGQENEGVVPFMASLELPYTVEV 389
Query 122 VPFSRSL 128
FS SL
Sbjct 390 RRFSPSL 396
> bbo:BBOV_III009670 17.m07839; dihydroorotase (EC:3.5.2.3); K01465
dihydroorotase [EC:3.5.2.3]
Length=351
Score = 91.7 bits (226), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 50/117 (42%), Positives = 66/117 (56%), Gaps = 14/117 (11%)
Query 1 LQVVREGHPQFFLGSDSAPHPRHKKDSYPPAAGVFTQPFLLAYLADTFARAGCLDKLQSF 60
LQ +R P+ FLGSDSAPH K+S P AG+FTQP+++ YLA F R C D L++F
Sbjct 235 LQAIRTRSPKIFLGSDSAPHTIAAKNSDKPPAGIFTQPYIMGYLATIFKRLDCEDYLEAF 294
Query 61 ACENAAAFFGLPKK--ELVEGEDCVVLEQTPCRIPNVVCGAEGTSTDVVPFLAGHEL 115
C N A F LP K E +E D ++ +P+ V G + PFLAG E+
Sbjct 295 VCRNGAEFLDLPAKNIEYMEITDETII------VPDTVGGV------LKPFLAGEEI 339
> pfa:PF14_0697 dihydroorotase, putative; K01465 dihydroorotase
[EC:3.5.2.3]
Length=358
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 49/124 (39%), Positives = 71/124 (57%), Gaps = 14/124 (11%)
Query 2 QVVREGHPQFFLGSDSAPHPR--HKKDSYPPAAGVFTQPFLLAYLADTFARAGCLDKLQS 59
V+++ P+ FLGSDSAPH + +K Y P G++TQPFL+ Y+A + LDK+++
Sbjct 243 DVIKDDFPRVFLGSDSAPHYKVMKRKPYYKP--GIYTQPFLINYVAHILNKFDALDKMEN 300
Query 60 FACENAAAFFGL-PKKELVEGEDCVVLEQTPCRIPNVVCGAEGTSTDVVPFLAGHELGYT 118
F +NA+ F L KK+L + CV E+ P ++P G VVPFLAG L Y
Sbjct 301 FTSKNASLFLNLAEKKKLAKYYICV--EKHPFKLPREYNG-------VVPFLAGKTLDYD 351
Query 119 LKVV 122
+ V
Sbjct 352 IHYV 355
> ath:AT4G22930 PYR4; PYR4 (PYRIMIDIN 4); dihydroorotase/ hydrolase/
hydrolase, acting on carbon-nitrogen (but not peptide)
bonds, in cyclic amides (EC:3.5.2.3); K01465 dihydroorotase
[EC:3.5.2.3]
Length=377
Score = 81.3 bits (199), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 42/117 (35%), Positives = 66/117 (56%), Gaps = 8/117 (6%)
Query 1 LQVVREGHPQFFLGSDSAPHPRHKKDSYPPAAGVFTQPFLLAYLADTFARAGCLDKLQSF 60
++ V G +FFLG+DSAPH R +K+S AG+++ P L+ A F AG LDKL++F
Sbjct 265 VKAVTSGSKKFFLGTDSAPHERSRKESSCGCAGIYSAPIALSLYAKVFDEAGALDKLEAF 324
Query 61 ACENAAAFFGLPKKELVEGEDCVVLEQTPCRIPNVVCGAEGTSTDVVPFLAGHELGY 117
N F+GLP+ + L+++P ++P+V G ++VP AG L +
Sbjct 325 TSFNGPDFYGLPR-----NSSKITLKKSPWKVPDVFNFPFG---EIVPMFAGETLQW 373
> sce:YLR420W URA4; Dihydroorotase, catalyzes the third enzymatic
step in the de novo biosynthesis of pyrimidines, converting
carbamoyl-L-aspartate into dihydroorotate (EC:3.5.2.3);
K01465 dihydroorotase [EC:3.5.2.3]
Length=364
Score = 74.3 bits (181), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 37/118 (31%), Positives = 64/118 (54%), Gaps = 4/118 (3%)
Query 7 GHPQFFLGSDSAPHPRHKKDSYPPA-AGVFTQPFLLAYLADTFARAGCLDKLQSFACENA 65
G P FF GSDSAPHP K +Y AGV++Q F + Y+A F L+ L+ F +
Sbjct 249 GKPYFFFGSDSAPHPVQNKANYEGVCAGVYSQSFAIPYIAQVFEEQNALENLKGFVSDFG 308
Query 66 AAFFGLPKKELVEGEDCVVLEQTPCRIPNVVCGAEGTSTDVVPFLAGHELGYTLKVVP 123
+F+ + E+ + ++ ++ IP V+ ++G ++PF AG +L ++++ P
Sbjct 309 ISFYEVKDSEVASSDKAILFKKEQV-IPQVI--SDGKDISIIPFKAGDKLSWSVRWEP 363
> eco:b1062 pyrC, ECK1047, JW1049; dihydro-orotase (EC:3.5.2.3);
K01465 dihydroorotase [EC:3.5.2.3]
Length=348
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 41/123 (33%), Positives = 55/123 (44%), Gaps = 16/123 (13%)
Query 2 QVVREGHPQFFLGSDSAPHPRHKKDSYPPAAGVFTQPFLLAYLADTFARAGCLDKLQSFA 61
++V G + FLG+DSAPH RH+K+S AG F P L A F L ++F
Sbjct 237 ELVASGFNRVFLGTDSAPHARHRKESSCGCAGCFNAPTALGSYATVFEEMNALQHFEAFC 296
Query 62 CENAAAFFGLPKK----ELVEGEDCVVLEQTPCRIPNVVCGAEGTSTDVVPFLAGHELGY 117
N F+GLP ELV E V T +VPFLAG + +
Sbjct 297 SVNGPQFYGLPVNDTFIELVREEQ------------QVAESIALTDDTLVPFLAGETVRW 344
Query 118 TLK 120
++K
Sbjct 345 SVK 347
> tpv:TP04_0882 dihydroorotase (EC:3.5.2.3); K01465 dihydroorotase
[EC:3.5.2.3]
Length=295
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 14/24 (58%), Positives = 20/24 (83%), Gaps = 0/24 (0%)
Query 1 LQVVREGHPQFFLGSDSAPHPRHK 24
L+ ++ GHP+FFLGSDSAPH ++
Sbjct 241 LETLKSGHPRFFLGSDSAPHTQYN 264
> mmu:327762 Dna2, Dna2l, E130315B21Rik, KIAA0083; DNA replication
helicase 2 homolog (yeast) (EC:3.6.4.12); K10742 DNA replication
ATP-dependent helicase Dna2 [EC:3.6.4.12]
Length=1062
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 24/51 (47%), Gaps = 3/51 (5%)
Query 3 VVREGHPQFFLGSDSAPHPRHKKDSYPPAAGVFTQPFLLAYLADTFARAGC 53
V+ +P FL +D P P ++ T+ L+ +L TF +AGC
Sbjct 886 VLEPDNPVCFLNTDKVPAPEQVENG---GVSNVTEARLIVFLTSTFIKAGC 933
> bbo:BBOV_III006600 17.m07584; hypothetical protein
Length=728
Score = 28.5 bits (62), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 10/21 (47%), Positives = 15/21 (71%), Gaps = 0/21 (0%)
Query 76 LVEGEDCVVLEQTPCRIPNVV 96
++E DCV + Q P R+PNV+
Sbjct 276 IIEFNDCVEVHQLPPRVPNVI 296
Lambda K H
0.322 0.138 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2099897216
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40