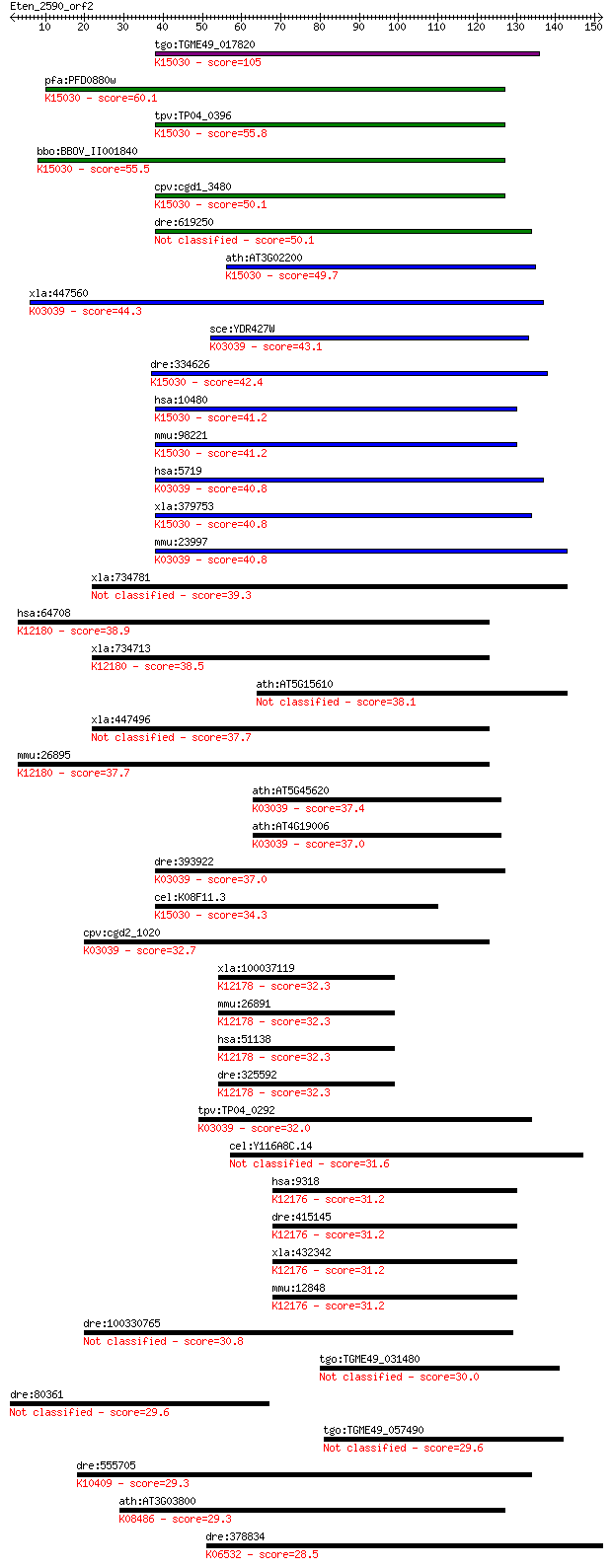
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2590_orf2
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_017820 proteasome PCI domain-containing protein ; K... 105 7e-23
pfa:PFD0880w proteasome regulatory component, putative; K15030... 60.1 2e-09
tpv:TP04_0396 hypothetical protein; K15030 translation initiat... 55.8 5e-08
bbo:BBOV_II001840 18.m06140; PCI domain containing protein; K1... 55.5 6e-08
cpv:cgd1_3480 proteasome regulatory complex component with a P... 50.1 3e-06
dre:619250 MGC114126; zgc:114126 50.1 3e-06
ath:AT3G02200 proteasome family protein; K15030 translation in... 49.7 3e-06
xla:447560 psmd13, MGC84231; proteasome (prosome, macropain) 2... 44.3 2e-04
sce:YDR427W RPN9; Non-ATPase regulatory subunit of the 26S pro... 43.1 4e-04
dre:334626 eif3m, GA17, fa16c10, pcid1, wu:fa16c10, wu:fc41f09... 42.4 6e-04
hsa:10480 EIF3M, B5, FLJ29030, PCID1, hfl-B5; eukaryotic trans... 41.2 0.001
mmu:98221 Eif3m, Ga17, MGC118449, Pcid1; eukaryotic translatio... 41.2 0.001
hsa:5719 PSMD13, HSPC027, Rpn9, S11, p40.5; proteasome (prosom... 40.8 0.001
xla:379753 eif3m, MGC52639, ga17, pcid1, tango7; eukaryotic tr... 40.8 0.002
mmu:23997 Psmd13, S11; proteasome (prosome, macropain) 26S sub... 40.8 0.002
xla:734781 hypothetical protein MGC131019 39.3 0.005
hsa:64708 COPS7B, CSN7B, FLJ12612, MGC111077; COP9 constitutiv... 38.9 0.006
xla:734713 cops7b, MGC114836; COP9 constitutive photomorphogen... 38.5 0.009
ath:AT5G15610 proteasome family protein 38.1 0.012
xla:447496 MGC81975 protein 37.7 0.013
mmu:26895 Cops7b, D1Wsu66e, E130114M23; COP9 (constitutive pho... 37.7 0.014
ath:AT5G45620 26S proteasome regulatory subunit, putative (RPN... 37.4 0.018
ath:AT4G19006 26S proteasome regulatory subunit, putative (RPN... 37.0 0.022
dre:393922 psmd13, MGC56377, zgc:56377, zgc:77482; proteasome ... 37.0 0.024
cel:K08F11.3 cif-1; COP9/Signalosome and eIF3 complex shared s... 34.3 0.15
cpv:cgd2_1020 proteasome regulatory subunit Rpn9, PINT domain ... 32.7 0.44
xla:100037119 cops4, COS41.8, SGN4; COP9 constitutive photomor... 32.3 0.56
mmu:26891 Cops4, AW208976, D5Ertd774e, MGC151177, MGC151203; C... 32.3 0.58
hsa:51138 COPS4, CSN4, MGC10899, MGC15160; COP9 constitutive p... 32.3 0.58
dre:325592 cops4, fc90c08, wu:fc90c08, zgc:77137; COP9 constit... 32.3 0.62
tpv:TP04_0292 26S proteasome regulatory subunit; K03039 26S pr... 32.0 0.70
cel:Y116A8C.14 rom-3; RhOMboid (Drosophila) related family mem... 31.6 0.96
hsa:9318 COPS2, ALIEN, CSN2, SGN2, TRIP15; COP9 constitutive p... 31.2 1.2
dre:415145 cops2, wu:fc05f05, wu:fc06g10, wu:fc60f04, wu:fl94f... 31.2 1.3
xla:432342 cops2, csn2; COP9 constitutive photomorphogenic hom... 31.2 1.3
mmu:12848 Cops2, AI315723, C85265, Csn2, Sgn2, Trip15, alien-l... 31.2 1.3
dre:100330765 serine/threonine kinase 31-like 30.8 1.9
tgo:TGME49_031480 translational activator, putative 30.0 2.8
dre:80361 or115-10, MGC123156, or9.3, zgc:123156; odorant rece... 29.6 3.5
tgo:TGME49_057490 prefoldin subunit 3, putative 29.6 3.8
dre:555705 zgc:158666; K10409 dynein intermediate chain 1, axo... 29.3 4.4
ath:AT3G03800 SYP131; SYP131 (SYNTAXIN OF PLANTS 131); SNAP re... 29.3 5.5
dre:378834 prom1b, proml2; prominin 1 b; K06532 prominin 28.5 8.1
> tgo:TGME49_017820 proteasome PCI domain-containing protein ;
K15030 translation initiation factor 3 subunit M
Length=463
Score = 105 bits (261), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 50/99 (50%), Positives = 75/99 (75%), Gaps = 1/99 (1%)
Query 38 KLQLLWVASTLQ-RRRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHV 96
K++LL VAS + R++ S + + D QLSEA AE++ VQAIGQG+V+AK+DQ+ +++HV
Sbjct 331 KIRLLAVASLVHGRKKEVSIRAIGDALQLSEAGAEEVAVQAIGQGIVDAKIDQLARVLHV 390
Query 97 RTAIQREFGRAQWEEVLGQLEHWASAVRTLRQAKESVKH 135
R+ +QREFGR QWEE+L +++HW+ VR L +SVK+
Sbjct 391 RSTMQREFGRQQWEELLERIDHWSEGVRALMGCMQSVKN 429
> pfa:PFD0880w proteasome regulatory component, putative; K15030
translation initiation factor 3 subunit M
Length=429
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/123 (28%), Positives = 65/123 (52%), Gaps = 6/123 (4%)
Query 10 IFCKSSANRALS-SSKSSSPSTPKCSTN-----TKLQLLWVASTLQRRRLFSFKQLSDFS 63
IF K + N L +K S K + + +K+ LL + S ++ + + +S
Sbjct 298 IFYKYTINEFLVFKNKHGSDFFNKYNIDIQTCESKIYLLSIISLFNDHKVQNIQFISKQL 357
Query 64 QLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASAV 123
+S E ++V AIG G+++AK+DQIN+ VH++T I R F W+++ Q+ + + V
Sbjct 358 NISVVQIENILVAAIGSGVIDAKIDQINQTVHMKTTILRNFDDENWKQLNNQITKYINNV 417
Query 124 RTL 126
+ +
Sbjct 418 QKI 420
> tpv:TP04_0396 hypothetical protein; K15030 translation initiation
factor 3 subunit M
Length=450
Score = 55.8 bits (133), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 51/89 (57%), Gaps = 0/89 (0%)
Query 38 KLQLLWVASTLQRRRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVR 97
KL LL +++ Q++ + + QL AEQ++V AI +G++EA +DQ +K V +
Sbjct 350 KLTLLTISTMCQQQSEIPIEMIEKNLQLPPEEAEQMIVNAINKGVMEALIDQNSKKVIIN 409
Query 98 TAIQREFGRAQWEEVLGQLEHWASAVRTL 126
+ REFG + +++ L+ W + + TL
Sbjct 410 HVVHREFGNEELKQLYNNLKQWRNCINTL 438
> bbo:BBOV_II001840 18.m06140; PCI domain containing protein;
K15030 translation initiation factor 3 subunit M
Length=311
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 65/119 (54%), Gaps = 5/119 (4%)
Query 8 FSIFCKSSANRALSSSKSSSPSTPKCSTNTKLQLLWVASTLQRRRLFSFKQLSDFSQLSE 67
F +F + AL++ SP T + KL+LL +AS Q ++ + +LS+
Sbjct 190 FDVFLSKHGDSALAAI-GISPETVR----AKLKLLTIASICQNEPEVPIARIQECLKLSK 244
Query 68 ATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASAVRTL 126
+E+LVV AI +G+++ +DQ ++ V +R+ +QR+F + Q +++ L W S V L
Sbjct 245 DESEELVVTAITKGVLDGLIDQRSEKVIIRSVMQRQFRKEQLQQLHSNLLQWKSCVSNL 303
> cpv:cgd1_3480 proteasome regulatory complex component with a
PINT domain at the C-terminus ; K15030 translation initiation
factor 3 subunit M
Length=456
Score = 50.1 bits (118), Expect = 3e-06, Method: Composition-based stats.
Identities = 24/89 (26%), Positives = 47/89 (52%), Gaps = 0/89 (0%)
Query 38 KLQLLWVASTLQRRRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVR 97
KLQLL +++ + + +L +LS + VV AI GL++ + + + V++
Sbjct 342 KLQLLTISTLAKGKSSIKLDELEKEFRLSSFDTQDAVVNAISVGLIDGNISENSNTVNIN 401
Query 98 TAIQREFGRAQWEEVLGQLEHWASAVRTL 126
+R+FG+A+WE + +L W + +L
Sbjct 402 CVTKRQFGKAEWESLDKKLNQWMGHLTSL 430
> dre:619250 MGC114126; zgc:114126
Length=370
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 47/96 (48%), Gaps = 0/96 (0%)
Query 38 KLQLLWVASTLQRRRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVR 97
K++LL + + +F ++ D Q+SE Q ++ + LV+A+V+ K + V
Sbjct 252 KMRLLTFMQMAEAKEELTFDEVQDQLQISEKDVHQFIIDVLKTKLVKARVNDKQKKILVS 311
Query 98 TAIQREFGRAQWEEVLGQLEHWASAVRTLRQAKESV 133
+ R FG+ QW + G L W + ++ L +V
Sbjct 312 ATMHRTFGKMQWAHLRGTLGSWETHLKNLSNGLTAV 347
> ath:AT3G02200 proteasome family protein; K15030 translation
initiation factor 3 subunit M
Length=417
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 45/79 (56%), Gaps = 0/79 (0%)
Query 56 FKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQ 115
+ + D Q++E E +V+AI L+E K+DQ+N+++ V + +REFG QW+ + +
Sbjct 317 YTSIKDTLQVNEQDVELWIVKAITAKLIECKMDQMNQVLIVSRSSEREFGTKQWQSLRTK 376
Query 116 LEHWASAVRTLRQAKESVK 134
L W + ++ ES K
Sbjct 377 LATWKDNISSIITTIESNK 395
> xla:447560 psmd13, MGC84231; proteasome (prosome, macropain)
26S subunit, non-ATPase, 13; K03039 26S proteasome regulatory
subunit N9
Length=378
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 37/144 (25%), Positives = 69/144 (47%), Gaps = 16/144 (11%)
Query 6 LFFSIFCKSSAN----RALSSSKSSSPSTPKCSTN-----TKLQLLWVAS-TLQR---RR 52
L +++ +S N RAL K++ P + N K+QLL + T R R
Sbjct 235 LIDTLYAFNSGNVETFRAL---KTAWGQQPDLAANEPLLLKKIQLLCLMEMTFTRPANHR 291
Query 53 LFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEV 112
+F++++ +Q+ E LV++A+ GL+ +D+++K VH+ R Q + +
Sbjct 292 QLTFEEIAKSAQVIVNDVELLVMKALSVGLLRGSIDEVDKRVHITWVQPRVLDLQQIKGM 351
Query 113 LGQLEHWASAVRTLRQAKESVKHS 136
+LEHW + V+++ E H
Sbjct 352 KDRLEHWCTDVKSMEMLVEHQAHD 375
> sce:YDR427W RPN9; Non-ATPase regulatory subunit of the 26S proteasome,
has similarity to putative proteasomal subunits in
other species; null mutant is temperature sensitive and exhibits
cell cycle and proteasome assembly defects; K03039 26S
proteasome regulatory subunit N9
Length=393
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 42/81 (51%), Gaps = 0/81 (0%)
Query 52 RLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEE 111
R+ SF+ +S + L + E LV++AI GL++ +DQ+N+LV + R Q +
Sbjct 306 RMLSFEDISKATHLPKDNVEHLVMRAISLGLLKGSIDQVNELVTISWVQPRIISGDQITK 365
Query 112 VLGQLEHWASAVRTLRQAKES 132
+ +L W V L + E+
Sbjct 366 MKDRLVEWNDQVEKLGKKMEA 386
> dre:334626 eif3m, GA17, fa16c10, pcid1, wu:fa16c10, wu:fc41f09,
zgc:63996; eukaryotic translation initiation factor 3, subunit
M (EC:3.6.5.3); K15030 translation initiation factor
3 subunit M
Length=375
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 24/101 (23%), Positives = 50/101 (49%), Gaps = 0/101 (0%)
Query 37 TKLQLLWVASTLQRRRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHV 96
+K++LL + SF+ + Q+ E V+ A+ +V +K+DQ + V V
Sbjct 275 SKMRLLTFMGMAVEMKEISFETMQQELQIGAEDVEAFVIDAVRTKMVYSKIDQTQRKVVV 334
Query 97 RTAIQREFGRAQWEEVLGQLEHWASAVRTLRQAKESVKHSA 137
+ R FG+ QW+++ L W + T++ + +++ +A
Sbjct 335 SHSTHRTFGKQQWQQLYDTLCSWKQNLSTVKSSLQTLSPTA 375
> hsa:10480 EIF3M, B5, FLJ29030, PCID1, hfl-B5; eukaryotic translation
initiation factor 3, subunit M (EC:3.6.5.3); K15030
translation initiation factor 3 subunit M
Length=374
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/92 (23%), Positives = 41/92 (44%), Gaps = 0/92 (0%)
Query 38 KLQLLWVASTLQRRRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVR 97
K++LL + SF + Q+ E V+ A+ +V K+DQ + V V
Sbjct 276 KMRLLTFMGMAVENKEISFDTMQQELQIGADDVEAFVIDAVRTKMVYCKIDQTQRKVVVS 335
Query 98 TAIQREFGRAQWEEVLGQLEHWASAVRTLRQA 129
+ R FG+ QW+++ L W + ++ +
Sbjct 336 HSTHRTFGKQQWQQLYDTLNAWKQNLNKVKNS 367
> mmu:98221 Eif3m, Ga17, MGC118449, Pcid1; eukaryotic translation
initiation factor 3, subunit M (EC:3.6.5.3); K15030 translation
initiation factor 3 subunit M
Length=374
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/92 (23%), Positives = 41/92 (44%), Gaps = 0/92 (0%)
Query 38 KLQLLWVASTLQRRRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVR 97
K++LL + SF + Q+ E V+ A+ +V K+DQ + V V
Sbjct 276 KMRLLTFMGMAVENKEISFDTMQQELQIGADDVEAFVIDAVRTKMVYCKIDQTQRKVVVS 335
Query 98 TAIQREFGRAQWEEVLGQLEHWASAVRTLRQA 129
+ R FG+ QW+++ L W + ++ +
Sbjct 336 HSTHRTFGKQQWQQLYDTLNAWKQNLNKVKNS 367
> hsa:5719 PSMD13, HSPC027, Rpn9, S11, p40.5; proteasome (prosome,
macropain) 26S subunit, non-ATPase, 13; K03039 26S proteasome
regulatory subunit N9
Length=376
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/103 (26%), Positives = 54/103 (52%), Gaps = 4/103 (3%)
Query 38 KLQLLWVAS-TLQR---RRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKL 93
K+QLL + T R R +F++++ ++++ E LV++A+ GLV+ +D+++K
Sbjct 271 KIQLLCLMEMTFTRPANHRQLTFEEIAKSAKITVNEVELLVMKALSVGLVKGSIDEVDKR 330
Query 94 VHVRTAIQREFGRAQWEEVLGQLEHWASAVRTLRQAKESVKHS 136
VH+ R Q + + +LE W + V+++ E H
Sbjct 331 VHMTWVQPRVLDLQQIKGMKDRLEFWCTDVKSMEMLVEHQAHD 373
> xla:379753 eif3m, MGC52639, ga17, pcid1, tango7; eukaryotic
translation initiation factor 3, subunit M (EC:3.6.5.3); K15030
translation initiation factor 3 subunit M
Length=374
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/96 (23%), Positives = 43/96 (44%), Gaps = 0/96 (0%)
Query 38 KLQLLWVASTLQRRRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVR 97
K++LL + SF + Q+ E ++ A+ +V K+DQ K V V
Sbjct 276 KMRLLTFMGMAVDNKEISFDTIQQELQMGADEVEAFIIDAVKTKMVYCKIDQTQKKVVVS 335
Query 98 TAIQREFGRAQWEEVLGQLEHWASAVRTLRQAKESV 133
+ R FG+ QW+++ L W + ++ + S+
Sbjct 336 HSTHRTFGKQQWQQLYDILNTWKLNLNKVKNSLYSI 371
> mmu:23997 Psmd13, S11; proteasome (prosome, macropain) 26S subunit,
non-ATPase, 13; K03039 26S proteasome regulatory subunit
N9
Length=376
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/109 (27%), Positives = 57/109 (52%), Gaps = 7/109 (6%)
Query 38 KLQLLWVAS-TLQR---RRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKL 93
K+QLL + T R R +F++++ ++++ E LV++A+ GLV +D+++K
Sbjct 271 KIQLLCLMEMTFTRPANHRQLTFEEIAKSAKITVNKVELLVMKALSVGLVRGSIDEVDKR 330
Query 94 VHVRTAIQREFGRAQWEEVLGQLEHWASAVRTLRQAKESVKHSAQPFET 142
VH+ R Q + + +LE W + V+++ V+H AQ T
Sbjct 331 VHMTWVQPRVLDLQQIKGMKDRLELWCTDVKSMEML---VEHQAQDILT 376
> xla:734781 hypothetical protein MGC131019
Length=216
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 31/133 (23%), Positives = 55/133 (41%), Gaps = 13/133 (9%)
Query 22 SSKSSSPSTPKCSTNTKLQLLWVASTLQRRRLFSFK-QLSDFSQLSEATAEQLVVQAIGQ 80
+SK S P N KL+ L + S R + + L D + E L+++AI
Sbjct 80 ASKDSLPELSAVQKN-KLKHLTIVSLASRMKCIPYSVLLKDLEMRNLRELEDLIIEAIYT 138
Query 81 GLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASAVRTL-----------RQA 129
+++ K+DQ N+++ V I R+ + ++ L+ W + Q
Sbjct 139 DIIQGKLDQRNQVLEVDFCIGRDIPKKDISSIVKTLQEWCDGCEAVLVGIEQQVVRANQY 198
Query 130 KESVKHSAQPFET 142
KE+ + Q ET
Sbjct 199 KETHNRTQQQIET 211
> hsa:64708 COPS7B, CSN7B, FLJ12612, MGC111077; COP9 constitutive
photomorphogenic homolog subunit 7B (Arabidopsis); K12180
COP9 signalosome complex subunit 7
Length=264
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 28/123 (22%), Positives = 52/123 (42%), Gaps = 12/123 (9%)
Query 3 LNILFFSIFCKSSANRALSSSKSSSPSTPKCST--NTKLQLLWVASTLQRRRLFSFK-QL 59
LN+ + + AN+ S P+ ST KL+ L + S R + + L
Sbjct 67 LNLFAYGTYPDYIANKE---------SLPELSTAQQNKLKHLTIVSLASRMKCIPYSVLL 117
Query 60 SDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHW 119
D + E L+++A+ +++ K+DQ N+L+ V I R+ + ++ L W
Sbjct 118 KDLEMRNLRELEDLIIEAVYTDIIQGKLDQRNQLLEVDFCIGRDIRKKDINNIVKTLHEW 177
Query 120 ASA 122
Sbjct 178 CDG 180
> xla:734713 cops7b, MGC114836; COP9 constitutive photomorphogenic
homolog subunit 7B; K12180 COP9 signalosome complex subunit
7
Length=264
Score = 38.5 bits (88), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 25/102 (24%), Positives = 46/102 (45%), Gaps = 2/102 (1%)
Query 22 SSKSSSPSTPKCSTNTKLQLLWVASTLQRRRLFSFK-QLSDFSQLSEATAEQLVVQAIGQ 80
+SK S P N KL+ L + S R + + L D + E L+++AI
Sbjct 80 ASKDSLPELSAVQKN-KLKHLTIVSLASRMKCIPYSVLLKDLEMRNLRELEDLIIEAIYT 138
Query 81 GLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASA 122
+++ K+DQ N+++ V I R+ + ++ L+ W
Sbjct 139 DIIQGKLDQRNQVLEVDFCIGRDIPKKDISSIVKTLQEWCDG 180
> ath:AT5G15610 proteasome family protein
Length=442
Score = 38.1 bits (87), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 29/108 (26%), Positives = 46/108 (42%), Gaps = 29/108 (26%)
Query 64 QLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRT------------------------- 98
Q+++ E VV+AI LV K+DQ+N++V VR
Sbjct 325 QVNDEEVELWVVKAITAKLVACKMDQMNQVVIVRQVSNLLLFRFICVNPKSHILVLHICS 384
Query 99 ----AIQREFGRAQWEEVLGQLEHWASAVRTLRQAKESVKHSAQPFET 142
+REFG+ QW+ + +L W VR + ES K + + +T
Sbjct 385 CISRCAEREFGQKQWQSLRTKLAAWRDNVRNVISTIESNKATEEGTQT 432
> xla:447496 MGC81975 protein
Length=264
Score = 37.7 bits (86), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 25/102 (24%), Positives = 45/102 (44%), Gaps = 2/102 (1%)
Query 22 SSKSSSPSTPKCSTNTKLQLLWVASTLQRRRLFSFK-QLSDFSQLSEATAEQLVVQAIGQ 80
+SK S P N KL+ L + S R + + L D + E L+++AI
Sbjct 80 ASKDSLPELSAVQKN-KLKHLTIVSLAARMKCIPYSVLLKDLEMRNLRELEDLIIEAIYT 138
Query 81 GLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASA 122
+++ K+DQ N ++ V I R+ + ++ L+ W
Sbjct 139 DIIQGKLDQRNHVLEVDFCIGRDIPKKDISSIVKTLQEWCDG 180
> mmu:26895 Cops7b, D1Wsu66e, E130114M23; COP9 (constitutive photomorphogenic)
homolog, subunit 7b (Arabidopsis thaliana);
K12180 COP9 signalosome complex subunit 7
Length=264
Score = 37.7 bits (86), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 28/121 (23%), Positives = 50/121 (41%), Gaps = 8/121 (6%)
Query 3 LNILFFSIFCKSSANRALSSSKSSSPSTPKCSTNTKLQLLWVASTLQRRRLFSFK-QLSD 61
LN+ + + AN K S P N KL+ L + S R + + L D
Sbjct 67 LNLFAYGTYPDYIAN------KESLPELSVAQQN-KLKHLTIVSLASRMKCIPYSVLLKD 119
Query 62 FSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWAS 121
+ E L+++A+ +++ K+DQ N+L+ V I R+ + ++ L W
Sbjct 120 LEMRNLRELEDLIIEAVYTDIIQGKLDQRNQLLEVDFCIGRDIRKKDINNIVKTLHEWCD 179
Query 122 A 122
Sbjct 180 G 180
> ath:AT5G45620 26S proteasome regulatory subunit, putative (RPN9);
K03039 26S proteasome regulatory subunit N9
Length=386
Score = 37.4 bits (85), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 34/63 (53%), Gaps = 0/63 (0%)
Query 63 SQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASA 122
++LS E L+++++ L+E +DQ+N ++V A R G Q + + QL+ W
Sbjct 309 TKLSIEDVEHLLMKSLSVHLIEGIIDQVNGTIYVSWAQPRVLGIPQIKALRDQLDSWVDK 368
Query 123 VRT 125
V T
Sbjct 369 VHT 371
> ath:AT4G19006 26S proteasome regulatory subunit, putative (RPN9);
K03039 26S proteasome regulatory subunit N9
Length=386
Score = 37.0 bits (84), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 34/63 (53%), Gaps = 0/63 (0%)
Query 63 SQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASA 122
++LS E L+++++ L+E +DQ+N V+V A R G Q + + QL+ W
Sbjct 309 TKLSIEDVEHLLMKSLSVHLIEGILDQVNGTVYVSWAQPRVLGIPQIKSLRDQLDSWVDK 368
Query 123 VRT 125
V T
Sbjct 369 VHT 371
> dre:393922 psmd13, MGC56377, zgc:56377, zgc:77482; proteasome
(prosome, macropain) 26S subunit, non-ATPase, 13; K03039 26S
proteasome regulatory subunit N9
Length=378
Score = 37.0 bits (84), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 48/93 (51%), Gaps = 4/93 (4%)
Query 38 KLQLLWVAS-TLQR---RRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKL 93
K+QLL V T R R SF++++ +++ E LV++A+ GL++ +D++ K
Sbjct 273 KIQLLCVMEMTFTRPANNRQLSFQEIAQSAKIQVNEVELLVMKALSVGLIKGSIDEVEKK 332
Query 94 VHVRTAIQREFGRAQWEEVLGQLEHWASAVRTL 126
VH+ R Q + + +L+ W V+ +
Sbjct 333 VHMTWVQPRVLDVQQIKGMKDRLDFWCGDVKNM 365
> cel:K08F11.3 cif-1; COP9/Signalosome and eIF3 complex shared
subunit family member (cif-1); K15030 translation initiation
factor 3 subunit M
Length=390
Score = 34.3 bits (77), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 38/73 (52%), Gaps = 1/73 (1%)
Query 38 KLQLLWVASTLQRRRLFSFKQLSD-FSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHV 96
K++LL + S + + S +L+ L++ T E+ V+ AI + K++++ + + V
Sbjct 287 KIRLLTLMSLAEEKNEISLDELAKQLDILADETLEEFVIDAIQVNAISGKINEMARTLIV 346
Query 97 RTAIQREFGRAQW 109
+ R FG QW
Sbjct 347 SSYQHRRFGTEQW 359
> cpv:cgd2_1020 proteasome regulatory subunit Rpn9, PINT domain
; K03039 26S proteasome regulatory subunit N9
Length=430
Score = 32.7 bits (73), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 19/106 (17%), Positives = 50/106 (47%), Gaps = 3/106 (2%)
Query 20 LSSSKSSSPSTPKCSTNTKLQLLWVASTLQRR---RLFSFKQLSDFSQLSEATAEQLVVQ 76
+S + S+P C T L + ++ R+ +F +++ ++ E LV++
Sbjct 307 VSKTPLSTPDARVCITKKTATLALMDLAFRKNKNERVLTFDEIAKHCRIGANEIELLVMK 366
Query 77 AIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASA 122
AI L++ +DQ ++ V + R + + + ++ ++++W +
Sbjct 367 AINMNLIKGIIDQASQTVEISWVHSRVLDKTRMKLLMDKIDNWIGS 412
> xla:100037119 cops4, COS41.8, SGN4; COP9 constitutive photomorphogenic
homolog subunit 4; K12178 COP9 signalosome complex
subunit 4
Length=406
Score = 32.3 bits (72), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 54 FSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRT 98
+F++L ++ A AE++ Q I +G + +DQI+ +VH T
Sbjct 319 ITFEELGALLEIPAAKAEKIASQMITEGRMNGFIDQIDGIVHFET 363
> mmu:26891 Cops4, AW208976, D5Ertd774e, MGC151177, MGC151203;
COP9 (constitutive photomorphogenic) homolog, subunit 4 (Arabidopsis
thaliana); K12178 COP9 signalosome complex subunit
4
Length=406
Score = 32.3 bits (72), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 54 FSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRT 98
+F++L ++ A AE++ Q I +G + +DQI+ +VH T
Sbjct 319 ITFEELGALLEIPAAKAEKIASQMITEGRMNGFIDQIDGIVHFET 363
> hsa:51138 COPS4, CSN4, MGC10899, MGC15160; COP9 constitutive
photomorphogenic homolog subunit 4 (Arabidopsis); K12178 COP9
signalosome complex subunit 4
Length=406
Score = 32.3 bits (72), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 54 FSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRT 98
+F++L ++ A AE++ Q I +G + +DQI+ +VH T
Sbjct 319 ITFEELGALLEIPAAKAEKIASQMITEGRMNGFIDQIDGIVHFET 363
> dre:325592 cops4, fc90c08, wu:fc90c08, zgc:77137; COP9 constitutive
photomorphogenic homolog subunit 4 (Arabidopsis); K12178
COP9 signalosome complex subunit 4
Length=406
Score = 32.3 bits (72), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 54 FSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRT 98
+F++L ++ A AE++ Q I +G + +DQI+ +VH T
Sbjct 319 ITFEELGALLEIPPAKAEKIASQMITEGRMNGFIDQIDGIVHFET 363
> tpv:TP04_0292 26S proteasome regulatory subunit; K03039 26S
proteasome regulatory subunit N9
Length=390
Score = 32.0 bits (71), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 18/85 (21%), Positives = 40/85 (47%), Gaps = 0/85 (0%)
Query 49 QRRRLFSFKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQ 108
++R SF+++ + ++ E +++A+G L++ +DQ + V R +
Sbjct 300 NKQRSLSFQEIVEHCKIQMDEVEPFILKALGCNLIQGHIDQTQGTIQVTWVQPRILDTNK 359
Query 109 WEEVLGQLEHWASAVRTLRQAKESV 133
E V +L+ W S+ L + E +
Sbjct 360 LELVRQKLKGWISSTNELVKGLEQL 384
> cel:Y116A8C.14 rom-3; RhOMboid (Drosophila) related family member
(rom-3)
Length=861
Score = 31.6 bits (70), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 26/98 (26%), Positives = 44/98 (44%), Gaps = 8/98 (8%)
Query 57 KQLSDFSQLSEATAEQLVVQAIGQGLVEAK----VDQINKLVHVRTAIQRE----FGRAQ 108
+Q+S F +++ E++ A +E + VD++NK +I+ E F
Sbjct 32 QQISHFIGIADTLEEKINWDAKRLRHIENRYKVIVDEVNKYAARHRSIEAETPLSFNEED 91
Query 109 WEEVLGQLEHWASAVRTLRQAKESVKHSAQPFETIPTP 146
E+ L WAS R Q K SV + ++T+P P
Sbjct 92 AEKGTRALPTWASIKRAFGQPKTSVGNYQILYDTLPQP 129
> hsa:9318 COPS2, ALIEN, CSN2, SGN2, TRIP15; COP9 constitutive
photomorphogenic homolog subunit 2 (Arabidopsis); K12176 COP9
signalosome complex subunit 2
Length=450
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 33/62 (53%), Gaps = 7/62 (11%)
Query 68 ATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASAVRTLR 127
A E L+VQ I + ++DQ+N+L+ + ++ G A++ L+ W + + +L
Sbjct 390 ADVESLLVQCILDNTIHGRIDQVNQLLELD---HQKRGGARY----TALDKWTNQLNSLN 442
Query 128 QA 129
QA
Sbjct 443 QA 444
> dre:415145 cops2, wu:fc05f05, wu:fc06g10, wu:fc60f04, wu:fl94f12,
zgc:86624; COP9 constitutive photomorphogenic homolog
subunit 2 (Arabidopsis); K12176 COP9 signalosome complex subunit
2
Length=443
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 33/62 (53%), Gaps = 7/62 (11%)
Query 68 ATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASAVRTLR 127
A E L+VQ I + ++DQ+N+L+ + ++ G A++ L+ W + + +L
Sbjct 383 ADVESLLVQCILDNTINGRIDQVNQLLELD---HQKRGGARY----TALDKWTNQLNSLN 435
Query 128 QA 129
QA
Sbjct 436 QA 437
> xla:432342 cops2, csn2; COP9 constitutive photomorphogenic homolog
subunit 2; K12176 COP9 signalosome complex subunit 2
Length=443
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 33/62 (53%), Gaps = 7/62 (11%)
Query 68 ATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASAVRTLR 127
A E L+VQ I + ++DQ+N+L+ + ++ G A++ L+ W + + +L
Sbjct 383 ADVESLLVQCILDNTINGRIDQVNQLLELD---HQKRGGARY----TALDKWTNQLNSLN 435
Query 128 QA 129
QA
Sbjct 436 QA 437
> mmu:12848 Cops2, AI315723, C85265, Csn2, Sgn2, Trip15, alien-like;
COP9 (constitutive photomorphogenic) homolog, subunit
2 (Arabidopsis thaliana); K12176 COP9 signalosome complex subunit
2
Length=443
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 33/62 (53%), Gaps = 7/62 (11%)
Query 68 ATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASAVRTLR 127
A E L+VQ I + ++DQ+N+L+ + ++ G A++ L+ W + + +L
Sbjct 383 ADVESLLVQCILDNTIHGRIDQVNQLLELD---HQKRGGARY----TALDKWTNQLNSLN 435
Query 128 QA 129
QA
Sbjct 436 QA 437
> dre:100330765 serine/threonine kinase 31-like
Length=931
Score = 30.8 bits (68), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 36/115 (31%), Positives = 53/115 (46%), Gaps = 9/115 (7%)
Query 20 LSSSKSSSPSTPKCSTNTKLQLLWVASTLQR-RRLFSFKQLSDFSQL-SEATAEQLVVQA 77
+ SS + + CS + + S LQR R + K + + +L E +A QL V
Sbjct 328 IKSSTTDADEQQLCSQDAEQVHNETESELQRMREVIDEKHMEEIKKLKEEGSAVQLHVLL 387
Query 78 IGQGLVEAKVD-QINKLVHVRTAIQREFGRAQWEEV---LGQLEHWASAVRTLRQ 128
+GQ L EAK++ Q K H +T +E G V + QL AVR LR+
Sbjct 388 LGQQLKEAKLELQTLKESHEKT---KEIGNNHPTTVANRISQLAKKVDAVRKLRE 439
> tgo:TGME49_031480 translational activator, putative
Length=3377
Score = 30.0 bits (66), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 31/64 (48%), Gaps = 5/64 (7%)
Query 80 QGLVEAKVDQINKLVHVRTAIQREFG---RAQWEEVLGQLEHWASAVRTLRQAKESVKHS 136
Q LV + ++ + HVR A R FG + EE L + W RTL+ ++ SV+ S
Sbjct 2179 QILVHLQTTLVDPIPHVRAAAARAFGTIAKGVGEEHLSDVLSW--LFRTLKTSESSVERS 2236
Query 137 AQPF 140
F
Sbjct 2237 GAAF 2240
> dre:80361 or115-10, MGC123156, or9.3, zgc:123156; odorant receptor,
family F, subfamily 115, member 10
Length=313
Score = 29.6 bits (65), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 33/66 (50%), Gaps = 5/66 (7%)
Query 1 IKLNILFFSIFCKSSANRALSSSKSSSPSTPKCSTNTKLQLLWVASTLQRRRLFSFKQLS 60
I L L +I C S N+A ++S + CST+ + L+ + S L L F +LS
Sbjct 214 IALTYLRIAIVCFKSKNKA-----TNSKAIKTCSTHLAVYLIMMVSGLTTITLHRFPELS 268
Query 61 DFSQLS 66
D +LS
Sbjct 269 DSRKLS 274
> tgo:TGME49_057490 prefoldin subunit 3, putative
Length=218
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 34/65 (52%), Gaps = 9/65 (13%)
Query 81 GLVEAKVDQI-NKLVHVRTAIQREFGRAQWEEVL---GQLEHWASAVRTLRQAKESVKHS 136
G VEA ++ K ++ AIQR Q E +L G +E SAV+ L+Q KE K +
Sbjct 60 GHVEAVARELLMKYRYMEQAIQR-----QSEGLLAKIGDMEEALSAVKRLKQQKEKAKDA 114
Query 137 AQPFE 141
A P E
Sbjct 115 ADPKE 119
> dre:555705 zgc:158666; K10409 dynein intermediate chain 1, axonemal
Length=660
Score = 29.3 bits (64), Expect = 4.4, Method: Composition-based stats.
Identities = 27/117 (23%), Positives = 49/117 (41%), Gaps = 7/117 (5%)
Query 18 RALSSSKSSSPSTPKCSTNTKLQLL-WVASTLQRRRLFSFKQLSDFSQLSEATAEQLVVQ 76
R +S P +T + ++ + LQRR KQ S F E +++ ++Q
Sbjct 179 REISCQTEPPPRANFSATANQWEIYDFYVEELQRREKSKEKQESQFPNKEEDRSKKKMIQ 238
Query 77 AIGQGLVEAKVDQINKLVHVRTAIQREFGRAQWEEVLGQLEHWASAVRTLRQAKESV 133
A EA+ D I++L I+R + ++EV ++ A R K ++
Sbjct 239 A------EAQSDDISQLAKPAEIIERMVNQNIFDEVAQDFMYFEDAADEFRGEKGTL 289
> ath:AT3G03800 SYP131; SYP131 (SYNTAXIN OF PLANTS 131); SNAP
receptor; K08486 syntaxin 1B/2/3
Length=306
Score = 29.3 bits (64), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 47/111 (42%), Gaps = 17/111 (15%)
Query 29 STPKCSTNTKLQLLWVASTLQRRRLFSFKQLSDFSQLSE-------ATAEQLVVQAIGQG 81
+ P C T + A+T+ ++ F K +S+F L + E+ V GQ
Sbjct 112 TKPGCGKGTGVDRTRTATTIAVKKKFKDK-ISEFQTLRQNIQQEYREVVERRVFTVTGQ- 169
Query 82 LVEAKVDQINKLVHVRTAIQ------REFGRAQWEEVLGQLEHWASAVRTL 126
A + I++L+ + Q RE GR Q + L +++ AVR L
Sbjct 170 --RADEEAIDRLIETGDSEQIFQKAIREQGRGQIMDTLAEIQERHDAVRDL 218
> dre:378834 prom1b, proml2; prominin 1 b; K06532 prominin
Length=851
Score = 28.5 bits (62), Expect = 8.1, Method: Composition-based stats.
Identities = 25/103 (24%), Positives = 43/103 (41%), Gaps = 7/103 (6%)
Query 51 RRLFS--FKQLSDFSQLSEATAEQLVVQAIGQGLVEAKVDQINKLVHVRTAIQREFGRAQ 108
RRL S + L F+ + A + L Q AK ++ L ++ T + +
Sbjct 203 RRLVSTNIRDLKAFANYTPAQIDYLTAQ-----YTTAKNKVLSDLDNIGTLLGGQIHNKL 257
Query 109 WEEVLGQLEHWASAVRTLRQAKESVKHSAQPFETIPTPVSTLQ 151
EV+ L+H +R+ KES++ + ET+ LQ
Sbjct 258 ESEVVPALDHALRMTAAMRETKESLESVSTSLETLQEGTGRLQ 300
Lambda K H
0.317 0.126 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3199347004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40