bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2593_orf3
Length=293
Score E
Sequences producing significant alignments: (Bits) Value
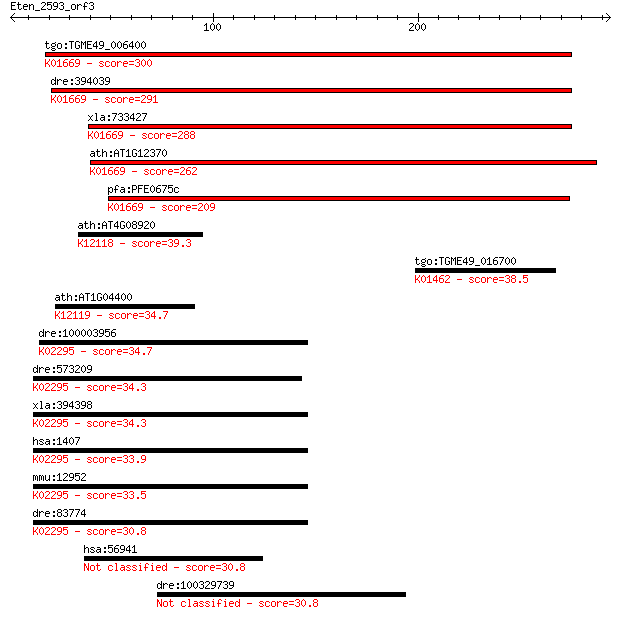
tgo:TGME49_006400 DNA photolyase, putative (EC:4.1.99.3); K016... 300 4e-81
dre:394039 MGC66475, phr; zgc:66475; K01669 deoxyribodipyrimid... 291 2e-78
xla:733427 phr; CPD photolyase-like; K01669 deoxyribodipyrimid... 288 1e-77
ath:AT1G12370 PHR1; PHR1 (PHOTOLYASE 1); DNA photolyase; K0166... 262 9e-70
pfa:PFE0675c deoxyribodipyrimidine photolyase (photoreactivati... 209 1e-53
ath:AT4G08920 CRY1; CRY1 (CRYPTOCHROME 1); ATP binding / blue ... 39.3 0.017
tgo:TGME49_016700 peptide deformylase, putative (EC:3.5.1.88);... 38.5 0.027
ath:AT1G04400 CRY2; CRY2 (CRYPTOCHROME 2); blue light photorec... 34.7 0.38
dre:100003956 cry1a, MGC153885, zcry1a, zgc:153885; cryptochro... 34.7 0.48
dre:573209 cry2a, MGC110521, zgc:110521; cryptochrome 2a; K022... 34.3 0.49
xla:394398 cry1, cry1-A, xCRY1; cryptochrome 1 (photolyase-lik... 34.3 0.64
hsa:1407 CRY1, PHLL1; cryptochrome 1 (photolyase-like); K02295... 33.9 0.74
mmu:12952 Cry1, AU020726, AU021000, Phll1; cryptochrome 1 (pho... 33.5 0.84
dre:83774 cry3, fe16d07, wu:fe16d07, zcry3; cryptochrome 3; K0... 30.8 6.3
hsa:56941 C3orf37, MGC111075; chromosome 3 open reading frame 37 30.8
dre:100329739 (6-4)-photolyase-like 30.8 6.4
> tgo:TGME49_006400 DNA photolyase, putative (EC:4.1.99.3); K01669
deoxyribodipyrimidine photo-lyase [EC:4.1.99.3]
Length=631
Score = 300 bits (769), Expect = 4e-81, Method: Compositional matrix adjust.
Identities = 148/261 (56%), Positives = 175/261 (67%), Gaps = 6/261 (2%)
Query 18 FPSD---ILEQLKADNRVQLRHCWRPGATAGLKAMKVFCTVPKLLAY-TSRNNPVLDSQS 73
FP D IL +LK D L W+PG A L+ + F T L Y SRN+P+ QS
Sbjct 263 FPLDEDRILSELKPDPPEPL-DSWKPGTKAALELLHSFATPKNLAEYGKSRNDPLAGVQS 321
Query 74 GLSPWLHFGQISAQRCLASVKDMGPATKIGATTAAARESYIEELVVRRELAENFTFYNSK 133
LSP++HFG IS QRCL V + +T T S+I+E+VVR +L++NF F+N
Sbjct 322 DLSPYIHFGHISVQRCLLEVSKLK-STAADKTLKEGVMSFIDEVVVRSQLSDNFVFFNPH 380
Query 134 YNQITGAPIWAQETLAAHKNDKREHLYALEQLEGSKTHDELWNAAQLQLVHTAKMHGFLR 193
Y+ I GAP+WAQ+TL H DKRE Y LE KT+DELWNAAQLQLV KMHG+LR
Sbjct 381 YDDIKGAPLWAQQTLNIHAKDKREPHYGFAALEAGKTYDELWNAAQLQLVRDGKMHGYLR 440
Query 194 MYWAKKILEWTPSADIALTTALFLNDKFSLDGTDPNGVVGCMWSVAGVHDQGWAERPIFG 253
MYWAKKILEWT S AL A+ LNDK+ LDGTDPNGV GCMWS+ GVHDQG+ ERP+FG
Sbjct 441 MYWAKKILEWTKSPQEALKIAIALNDKYHLDGTDPNGVTGCMWSICGVHDQGFKERPVFG 500
Query 254 KIRYMNLAGCMRKFSVDAFVR 274
KIRYMN GC RKF V AFVR
Sbjct 501 KIRYMNYPGCERKFDVKAFVR 521
> dre:394039 MGC66475, phr; zgc:66475; K01669 deoxyribodipyrimidine
photo-lyase [EC:4.1.99.3]
Length=516
Score = 291 bits (746), Expect = 2e-78, Method: Compositional matrix adjust.
Identities = 141/254 (55%), Positives = 180/254 (70%), Gaps = 7/254 (2%)
Query 21 DILEQLKADNRVQLRHCWRPGATAGLKAMKVFCTVPKLLAYTSRNNPVLDSQSGLSPWLH 80
++L L+ D+ V +PG T G+ ++ F + T RNNP D+ S LSPW+H
Sbjct 253 EVLSSLEVDHNVCEVEWAQPGTTGGMFMLESFIDQRLHIFATHRNNPNSDAVSHLSPWIH 312
Query 81 FGQISAQRCLASVKDMGPATKIGATTAAARESYIEELVVRRELAENFTFYNSKYNQITGA 140
GQ+SAQR + VK K A+ + A S+IEE+VVRRELA+NF FYN Y+ I+GA
Sbjct 313 AGQLSAQRVVMQVK-----RKKNASESVA--SFIEEIVVRRELADNFCFYNQNYDNISGA 365
Query 141 PIWAQETLAAHKNDKREHLYALEQLEGSKTHDELWNAAQLQLVHTAKMHGFLRMYWAKKI 200
WA++TL H D+R++LY ++LE ++THD+LWNAAQ QL+ KMHGFLRMYWAKKI
Sbjct 366 YDWAKKTLQEHAKDRRQYLYTKKELESAETHDQLWNAAQRQLLLEGKMHGFLRMYWAKKI 425
Query 201 LEWTPSADIALTTALFLNDKFSLDGTDPNGVVGCMWSVAGVHDQGWAERPIFGKIRYMNL 260
LEWT S + AL+ A++LND+ SLDG DPNG VGCMWS+ G+HDQGWAERPIFGKIRYMN
Sbjct 426 LEWTASPEEALSIAIYLNDRLSLDGCDPNGYVGCMWSICGIHDQGWAERPIFGKIRYMNY 485
Query 261 AGCMRKFSVDAFVR 274
AGC RKF V+ F R
Sbjct 486 AGCKRKFDVEQFER 499
> xla:733427 phr; CPD photolyase-like; K01669 deoxyribodipyrimidine
photo-lyase [EC:4.1.99.3]
Length=557
Score = 288 bits (737), Expect = 1e-77, Method: Compositional matrix adjust.
Identities = 134/236 (56%), Positives = 163/236 (69%), Gaps = 6/236 (2%)
Query 39 RPGATAGLKAMKVFCTVPKLLAYTSRNNPVLDSQSGLSPWLHFGQISAQRCLASVKDMGP 98
+PG AG ++ F + + RNNP ++ S LSPW HFGQ+S QR + V+
Sbjct 319 KPGTNAGFNVLQSFISERLKHFNSDRNNPNQNALSNLSPWFHFGQLSVQRAILEVQ---- 374
Query 99 ATKIGATTAAARESYIEELVVRRELAENFTFYNSKYNQITGAPIWAQETLAAHKNDKREH 158
K + + +S++EE VVRRELA+NF FYN Y++I GA WA+ TL H DKR H
Sbjct 375 --KYRSKFKESVDSFVEEAVVRRELADNFCFYNKNYDKIEGAYDWAKNTLKDHAKDKRTH 432
Query 159 LYALEQLEGSKTHDELWNAAQLQLVHTAKMHGFLRMYWAKKILEWTPSADIALTTALFLN 218
LY LE+LE KTHD LWNAAQLQ+VH KMHGFLRMYWAKKILEWT S + AL +L+LN
Sbjct 433 LYTLEKLEAGKTHDPLWNAAQLQMVHEGKMHGFLRMYWAKKILEWTSSPEEALHFSLYLN 492
Query 219 DKFSLDGTDPNGVVGCMWSVAGVHDQGWAERPIFGKIRYMNLAGCMRKFSVDAFVR 274
D++ LDG DPNG VGCMWS+ G+HDQGWAER +FGKIRYMN GC RKF V F R
Sbjct 493 DRYELDGRDPNGYVGCMWSICGIHDQGWAERAVFGKIRYMNYQGCKRKFDVAQFER 548
> ath:AT1G12370 PHR1; PHR1 (PHOTOLYASE 1); DNA photolyase; K01669
deoxyribodipyrimidine photo-lyase [EC:4.1.99.3]
Length=490
Score = 262 bits (670), Expect = 9e-70, Method: Compositional matrix adjust.
Identities = 127/252 (50%), Positives = 175/252 (69%), Gaps = 17/252 (6%)
Query 40 PGATAGLKAM---KVFCTVPKLLAY-TSRNNPVL-DSQSGLSPWLHFGQISAQRCLASVK 94
PG AG++ + K +L Y T RNNP+ + SGLSP+LHFGQ+SAQRC
Sbjct 232 PGEDAGIEVLMGNKDGFLTKRLKNYSTDRNNPIKPKALSGLSPYLHFGQVSAQRCALE-- 289
Query 95 DMGPATKIGATTAAARESYIEELVVRRELAENFTFYNSKYNQITGAPIWAQETLAAHKND 154
A K+ +T+ A ++++EEL+VRREL++NF +Y Y+ + GA WA+++L H +D
Sbjct 290 ----ARKVRSTSPQAVDTFLEELIVRRELSDNFCYYQPHYDSLKGAWEWARKSLMDHASD 345
Query 155 KREHLYALEQLEGSKTHDELWNAAQLQLVHTAKMHGFLRMYWAKKILEWTPSADIALTTA 214
KREH+Y+LEQLE T D L ++V+ KMHGF+RMYWAKKILEWT + AL+ +
Sbjct 346 KREHIYSLEQLEKGLTADPL------EMVYQGKMHGFMRMYWAKKILEWTKGPEEALSIS 399
Query 215 LFLNDKFSLDGTDPNGVVGCMWSVAGVHDQGWAERPIFGKIRYMNLAGCMRKFSVDAFVR 274
++LN+K+ +DG DP+G VGCMWS+ GVHDQGW ERP+FGKIRYMN AGC RKF+VD+++
Sbjct 400 IYLNNKYEIDGRDPSGYVGCMWSICGVHDQGWKERPVFGKIRYMNYAGCKRKFNVDSYIS 459
Query 275 CVRQIVGRHAKK 286
V+ +V KK
Sbjct 460 YVKSLVSVTKKK 471
> pfa:PFE0675c deoxyribodipyrimidine photolyase (photoreactivating
enzyme, DNA photolyase), putative (EC:4.1.99.3); K01669
deoxyribodipyrimidine photo-lyase [EC:4.1.99.3]
Length=1113
Score = 209 bits (532), Expect = 1e-53, Method: Composition-based stats.
Identities = 94/226 (41%), Positives = 145/226 (64%), Gaps = 3/226 (1%)
Query 49 MKVFCTVPKLLAYT-SRNNPVLDSQSGLSPWLHFGQISAQRCLASVKDMGPATKIGATTA 107
++ FC+ KL Y+ +N+P + + L+P+++FG IS+QRC+ V + T
Sbjct 840 LQNFCS-KKLERYSLKKNDPNSEMINLLTPYINFGIISSQRCVLEVNKYANSYP-SINTI 897
Query 108 AARESYIEELVVRRELAENFTFYNSKYNQITGAPIWAQETLAAHKNDKREHLYALEQLEG 167
+ +E + EE+++++ELA+NF +YN Y+ G WA+++L H +DKRE+LY + +
Sbjct 898 SGKELFSEEMIMKKELADNFCYYNKNYDNFNGGKDWAKDSLKKHDSDKREYLYDFDDFKN 957
Query 168 SKTHDELWNAAQLQLVHTAKMHGFLRMYWAKKILEWTPSADIALTTALFLNDKFSLDGTD 227
+KTHD+LWN QLQL++ +H FLRMYW KKIL W+ ++ AL A+ LND FS+D
Sbjct 958 AKTHDDLWNCCQLQLINEGIIHEFLRMYWCKKILNWSGNSKTALKCAMKLNDDFSIDAKS 1017
Query 228 PNGVVGCMWSVAGVHDQGWAERPIFGKIRYMNLAGCMRKFSVDAFV 273
P+G V M S+ G+HDQ W ER +FGKIR MN C R F ++ ++
Sbjct 1018 PHGYVSIMSSIMGIHDQSWNERTVFGKIRSMNYNSCKRLFDINVYM 1063
> ath:AT4G08920 CRY1; CRY1 (CRYPTOCHROME 1); ATP binding / blue
light photoreceptor/ protein homodimerization/ protein kinase;
K12118 cryptochrome 1
Length=681
Score = 39.3 bits (90), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 34/63 (53%), Gaps = 4/63 (6%)
Query 34 LRHCWRPGATAGLKAMKVFCTVPKLLAYTSRNNPVLDS--QSGLSPWLHFGQISAQRCLA 91
L W PG + G KA+ F P LL Y S+N DS S LSP LHFG++S ++
Sbjct 209 LARAWSPGWSNGDKALTTFINGP-LLEY-SKNRRKADSATTSFLSPHLHFGEVSVRKVFH 266
Query 92 SVK 94
V+
Sbjct 267 LVR 269
> tgo:TGME49_016700 peptide deformylase, putative (EC:3.5.1.88);
K01462 peptide deformylase [EC:3.5.1.88]
Length=353
Score = 38.5 bits (88), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 40/71 (56%), Gaps = 6/71 (8%)
Query 199 KILEWTPSADIALTTA--LFLNDKF-SLDGTDPNGVVGCMWSVAGVHDQGWAERPIFGKI 255
+++ W P+ D+ ++ +FLN + SL G + V GC+ SV GV ERP+ ++
Sbjct 239 QMIVWNPTGDVRESSRERVFLNPRLLSLYGPLVSDVEGCL-SVPGVF--APVERPLHARV 295
Query 256 RYMNLAGCMRK 266
RY +L G R+
Sbjct 296 RYTSLEGIQRE 306
> ath:AT1G04400 CRY2; CRY2 (CRYPTOCHROME 2); blue light photoreceptor/
protein homodimerization; K12119 cryptochrome 2
Length=612
Score = 34.7 bits (78), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 37/75 (49%), Gaps = 8/75 (10%)
Query 23 LEQLKADNRVQ------LRHCWRPGATAGLKAMKVFCTVPKLLAYTSRNNPVL-DSQSGL 75
+E+L +N + L W PG + K + F +L+ Y + V+ +S S L
Sbjct 189 IEELGLENEAEKPSNALLTRAWSPGWSNADKLLNEFIE-KQLIDYAKNSKKVVGNSTSLL 247
Query 76 SPWLHFGQISAQRCL 90
SP+LHFG+IS +
Sbjct 248 SPYLHFGEISVRHVF 262
> dre:100003956 cry1a, MGC153885, zcry1a, zgc:153885; cryptochrome
1a; K02295 cryptochrome
Length=619
Score = 34.7 bits (78), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 33/135 (24%), Positives = 56/135 (41%), Gaps = 8/135 (5%)
Query 15 DARFPSDILEQLKADNRVQLRHCWRPGATAGLKAMKVFCTVPKLLAYTSRN----NPVLD 70
D +F LE+L D W G T L ++ +A R N +L
Sbjct 187 DEKFGVPSLEELGFDTEGLSSAVWPGGETEALTRLERHLERKAWVANFERPRMNANSLLA 246
Query 71 SQSGLSPWLHFGQISAQRCLASVKDMGPATKIGATTAAARESYIEELVVRRELAENFTFY 130
S +GLSP+L FG +S + + D+ K+ ++ Y + ++ RE
Sbjct 247 SPTGLSPYLRFGCLSCRLFYFKLTDL--YRKVKKNSSPPLSLYGQ--LLWREFFYTAATN 302
Query 131 NSKYNQITGAPIWAQ 145
N +++++ G PI Q
Sbjct 303 NPRFDKMEGNPICVQ 317
> dre:573209 cry2a, MGC110521, zgc:110521; cryptochrome 2a; K02295
cryptochrome
Length=655
Score = 34.3 bits (77), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 35/135 (25%), Positives = 57/135 (42%), Gaps = 10/135 (7%)
Query 12 HGEDARFPSDILEQLKADNRVQLRHCWRPGATAGLKAMKVFCTVPKLLAYTSRN----NP 67
HG+ PS LE+L D W G T L ++ +A R N
Sbjct 186 HGDKYGVPS--LEELGFDIEGLPSAVWPGGETEALTRIERHLERKAWVANFERPRMNANS 243
Query 68 VLDSQSGLSPWLHFGQISAQRCLASVKDMGPATKIGATTAAARESYIEELVVRRELAENF 127
+L S +GLSP+L FG +S + + D+ K+ T+ Y + ++ RE
Sbjct 244 LLASPTGLSPYLRFGCLSCRLFYFKLTDL--YRKVKKTSTPPLSLYGQ--LLWREFFYTA 299
Query 128 TFYNSKYNQITGAPI 142
N +++++ G PI
Sbjct 300 ATTNPRFDKMEGNPI 314
> xla:394398 cry1, cry1-A, xCRY1; cryptochrome 1 (photolyase-like);
K02295 cryptochrome
Length=616
Score = 34.3 bits (77), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 35/138 (25%), Positives = 57/138 (41%), Gaps = 10/138 (7%)
Query 12 HGEDARFPSDILEQLKADNRVQLRHCWRPGATAGLKAMKVFCTVPKLLAYTSRN----NP 67
H E PS LE+L D W G T L ++ +A R N
Sbjct 186 HDEKYGVPS--LEELGFDTEGLPSAVWPGGETEALTRLERHLERKAWVANFERPRMNANS 243
Query 68 VLDSQSGLSPWLHFGQISAQRCLASVKDMGPATKIGATTAAARESYIEELVVRRELAENF 127
+L S +GLSP+L FG +S + + D+ K+ ++ Y + ++ RE
Sbjct 244 LLASTTGLSPYLRFGCLSCRLFYFKLTDL--YKKVKKNSSPPLSLYGQ--LLWREFFYTA 299
Query 128 TFYNSKYNQITGAPIWAQ 145
N +++++ G PI Q
Sbjct 300 ATNNPRFDKMDGNPICVQ 317
> hsa:1407 CRY1, PHLL1; cryptochrome 1 (photolyase-like); K02295
cryptochrome
Length=586
Score = 33.9 bits (76), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 35/138 (25%), Positives = 57/138 (41%), Gaps = 10/138 (7%)
Query 12 HGEDARFPSDILEQLKADNRVQLRHCWRPGATAGLKAMKVFCTVPKLLAYTSRN----NP 67
H E PS LE+L D W G T L ++ +A R N
Sbjct 186 HDEKYGVPS--LEELGFDTDGLSSAVWPGGETEALTRLERHLERKAWVANFERPRMNANS 243
Query 68 VLDSQSGLSPWLHFGQISAQRCLASVKDMGPATKIGATTAAARESYIEELVVRRELAENF 127
+L S +GLSP+L FG +S + + D+ K+ ++ Y + ++ RE
Sbjct 244 LLASPTGLSPYLRFGCLSCRLFYFKLTDL--YKKVKKNSSPPLSLYGQ--LLWREFFYTA 299
Query 128 TFYNSKYNQITGAPIWAQ 145
N +++++ G PI Q
Sbjct 300 ATNNPRFDKMEGNPICVQ 317
> mmu:12952 Cry1, AU020726, AU021000, Phll1; cryptochrome 1 (photolyase-like);
K02295 cryptochrome
Length=606
Score = 33.5 bits (75), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 35/138 (25%), Positives = 57/138 (41%), Gaps = 10/138 (7%)
Query 12 HGEDARFPSDILEQLKADNRVQLRHCWRPGATAGLKAMKVFCTVPKLLAYTSRN----NP 67
H E PS LE+L D W G T L ++ +A R N
Sbjct 186 HDEKYGVPS--LEELGFDTDGLSSAVWPGGETEALTRLERHLERKAWVANFERPRMNANS 243
Query 68 VLDSQSGLSPWLHFGQISAQRCLASVKDMGPATKIGATTAAARESYIEELVVRRELAENF 127
+L S +GLSP+L FG +S + + D+ K+ ++ Y + ++ RE
Sbjct 244 LLASPTGLSPYLRFGCLSCRLFYFKLTDL--YKKVKKNSSPPLSLYGQ--LLWREFFYTA 299
Query 128 TFYNSKYNQITGAPIWAQ 145
N +++++ G PI Q
Sbjct 300 ATNNPRFDKMEGNPICVQ 317
> dre:83774 cry3, fe16d07, wu:fe16d07, zcry3; cryptochrome 3;
K02295 cryptochrome
Length=598
Score = 30.8 bits (68), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 32/138 (23%), Positives = 57/138 (41%), Gaps = 10/138 (7%)
Query 12 HGEDARFPSDILEQLKADNRVQLRHCWRPGATAGLKAMKVFCTVPKLLAYTSR----NNP 67
H E PS LE+L + H W+ G T L+ + +A R
Sbjct 186 HDEHYGVPS--LEELGFRTQGDSLHVWKGGETEALERLNKHLDRKAWVANFERPRISGQS 243
Query 68 VLDSQSGLSPWLHFGQISAQRCLASVKDMGPATKIGATTAAARESYIEELVVRRELAENF 127
+ S +GLSP+L FG +S + +++D+ K+ ++ + + ++ RE
Sbjct 244 LFPSPTGLSPYLRFGCLSCRVFYYNLRDL--FMKLRRRSSPPLSLFGQ--LLWREFFYTA 299
Query 128 TFYNSKYNQITGAPIWAQ 145
N ++ + G PI Q
Sbjct 300 GTNNPNFDHMEGNPICVQ 317
> hsa:56941 C3orf37, MGC111075; chromosome 3 open reading frame
37
Length=354
Score = 30.8 bits (68), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 24/97 (24%), Positives = 39/97 (40%), Gaps = 10/97 (10%)
Query 37 CWRPGATAGLKAMKVFCTVPKLLAYTS---RNNPVLDSQSGLSPWLHFGQISAQRCLASV 93
CW P + TV + R +LD + +S WL FG++S Q L +
Sbjct 182 CWEPPEGGDVLYSYTIITVDSCKGLSDIHHRMPAILDGEEAVSKWLDFGEVSTQEALKLI 241
Query 94 KDMGPAT--KIGATTAAARESYIE-----ELVVRREL 123
T + + +R + E +LVV++EL
Sbjct 242 HPTENITFHAVSSVVNNSRNNTPECLAPVDLVVKKEL 278
> dre:100329739 (6-4)-photolyase-like
Length=813
Score = 30.8 bits (68), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 57/126 (45%), Gaps = 28/126 (22%)
Query 73 SGLSPWLHFGQISAQRCLASVKDMGPATKIGATTAAARESYIEELVVRRELAENFTFYNS 132
S LSP+LHFGQ+SA++ L AAR + + +R+LA + +
Sbjct 437 SCLSPYLHFGQLSARQVL----------------WAARGARCKSPKFQRKLA----WRDL 476
Query 133 KYNQITGAPIWAQETL-----AAHKNDKREHLYALEQLEGSKTHDELWNAAQLQLVHTAK 187
Y QI+ P E+L A + HL A ++ +T L +AA QL T
Sbjct 477 AYWQISLFPDLPWESLRPPYKALRWSSDHAHLKAWQR---GRTGYPLVDAAMRQLWQTGW 533
Query 188 MHGFLR 193
M+ ++R
Sbjct 534 MNNYMR 539
Lambda K H
0.320 0.133 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 11520258316
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40