bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2679_orf3
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
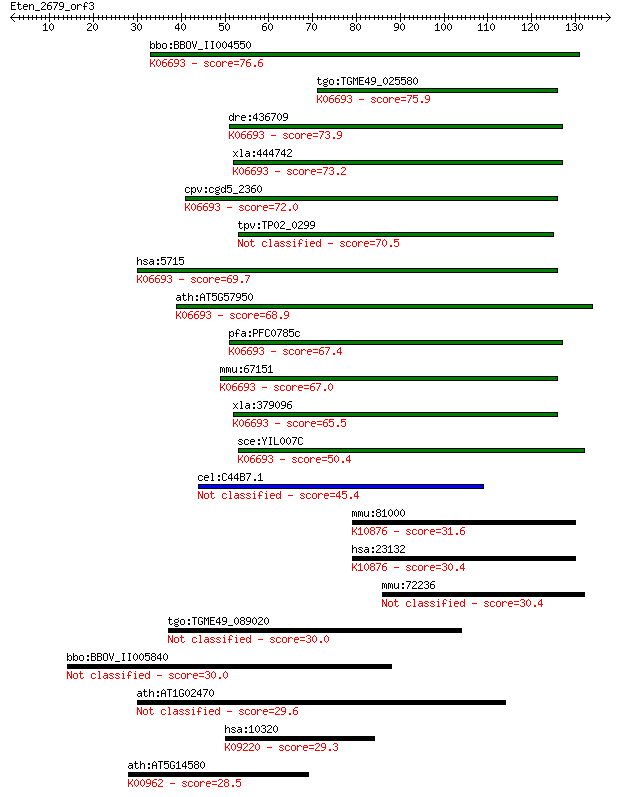
bbo:BBOV_II004550 18.m06379; hypothetical protein; K06693 26S ... 76.6 2e-14
tgo:TGME49_025580 26S Proteasome non-ATPase regulatory subunit... 75.9 3e-14
dre:436709 psmd9, wu:fb30g07, zgc:92643; proteasome (prosome, ... 73.9 1e-13
xla:444742 psmd9, MGC84770; proteasome (prosome, macropain) 26... 73.2 2e-13
cpv:cgd5_2360 p27 like 26S proteasomal subunit with a PDZ doma... 72.0 5e-13
tpv:TP02_0299 hypothetical protein 70.5 2e-12
hsa:5715 PSMD9, MGC8644, Rpn4, p27; proteasome (prosome, macro... 69.7 3e-12
ath:AT5G57950 26S proteasome regulatory subunit, putative; K06... 68.9 4e-12
pfa:PFC0785c proteasome regulatory protein, putative; K06693 2... 67.4 1e-11
mmu:67151 Psmd9, 1500011J20Rik, 2610202L11Rik, Bridge-1, P27; ... 67.0 1e-11
xla:379096 MGC53232; similar to proteasome (prosome, macropain... 65.5 5e-11
sce:YIL007C NAS2; Nas2p; K06693 26S proteasome non-ATPase regu... 50.4 2e-06
cel:C44B7.1 hypothetical protein 45.4 5e-05
mmu:81000 Rad54l2, Arip4, D130058C05, G630026H09Rik, Srisnf2l;... 31.6 0.70
hsa:23132 RAD54L2, ARIP4, FLJ21396, FLJ22400, HSPC325, KIAA080... 30.4 1.5
mmu:72236 Tsnaxip1, 1700016K08Rik, MGC118240, TXI1; translin-a... 30.4 1.8
tgo:TGME49_089020 hypothetical protein 30.0 2.0
bbo:BBOV_II005840 18.m06484; hypothetical protein 30.0 2.3
ath:AT1G02470 hypothetical protein 29.6 2.7
hsa:10320 IKZF1, Hs.54452, IK1, IKAROS, LYF1, PRO0758, ZNFN1A1... 29.3 4.3
ath:AT5G14580 polyribonucleotide nucleotidyltransferase, putat... 28.5 7.1
> bbo:BBOV_II004550 18.m06379; hypothetical protein; K06693 26S
proteasome non-ATPase regulatory subunit 9
Length=100
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 40/100 (40%), Positives = 62/100 (62%), Gaps = 4/100 (4%)
Query 33 MAASAGASSLAAQAEAVYRERQQIEEKMEALASFLTAEGMP--GLKGPLVDPEGFPRADI 90
M + + A+ + + R+ IE +MEAL ++L +E G+KGPLVD E +PR+DI
Sbjct 1 MEQKSANNDTIARINELSKIRKDIEVEMEALLNYLNSEECKHVGMKGPLVDEEQYPRSDI 60
Query 91 DVHAVLQARQQLACLKTDHREVQQRLEETLLLLHAANAKD 130
D+ AV AR ++ CL TD++E++ +L + L LH N KD
Sbjct 61 DICAVRNARHRINCLHTDYKELEDKLAQALHELH--NKKD 98
> tgo:TGME49_025580 26S Proteasome non-ATPase regulatory subunit
9, putative ; K06693 26S proteasome non-ATPase regulatory
subunit 9
Length=219
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 34/55 (61%), Positives = 44/55 (80%), Gaps = 0/55 (0%)
Query 71 GMPGLKGPLVDPEGFPRADIDVHAVLQARQQLACLKTDHREVQQRLEETLLLLHA 125
GMPGL G LVD EGFPRADID++A+ AR +LA LKTD++EV+ ++E+ L LHA
Sbjct 8 GMPGLTGGLVDDEGFPRADIDIYAIRGARNRLAILKTDYKEVRSQIEKELFALHA 62
> dre:436709 psmd9, wu:fb30g07, zgc:92643; proteasome (prosome,
macropain) 26S subunit, non-ATPase, 9; K06693 26S proteasome
non-ATPase regulatory subunit 9
Length=211
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 32/76 (42%), Positives = 49/76 (64%), Gaps = 0/76 (0%)
Query 51 RERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRADIDVHAVLQARQQLACLKTDHR 110
+ + IEE+++A L ++ G+ GPLVD EGFPRAD+D++ V AR ++CL+ DH+
Sbjct 24 KRKDDIEEQIKAYYDMLESQAGVGMDGPLVDVEGFPRADVDLYKVRTARHSISCLQNDHK 83
Query 111 EVQQRLEETLLLLHAA 126
+ +EE L LHA
Sbjct 84 AIMVEIEEALHKLHAT 99
> xla:444742 psmd9, MGC84770; proteasome (prosome, macropain)
26S subunit, non-ATPase, 9; K06693 26S proteasome non-ATPase
regulatory subunit 9
Length=208
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 52/75 (69%), Gaps = 0/75 (0%)
Query 52 ERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRADIDVHAVLQARQQLACLKTDHRE 111
++ ++E +++AL L + G+ GPLVD EG+PRAD+D++ V AR + CL+ DH+
Sbjct 23 KKDEMEAEIKALYDLLQDQKGIGMDGPLVDREGYPRADVDIYQVRTARHNIICLQNDHKA 82
Query 112 VQQRLEETLLLLHAA 126
+ +++E++L +LHA
Sbjct 83 IMKKIEQSLHILHAG 97
> cpv:cgd5_2360 p27 like 26S proteasomal subunit with a PDZ domain
; K06693 26S proteasome non-ATPase regulatory subunit 9
Length=249
Score = 72.0 bits (175), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 55/86 (63%), Gaps = 1/86 (1%)
Query 41 SLAAQAEAVYRERQQIEEKMEALASFLTAEG-MPGLKGPLVDPEGFPRADIDVHAVLQAR 99
++ E + + + +IE+++ L FL + G G+ G LVD EGFPR+DID++AV +AR
Sbjct 8 NINVNMEELAKRKGEIEKEVSELTEFLNSCGPDVGISGKLVDSEGFPRSDIDIYAVRRAR 67
Query 100 QQLACLKTDHREVQQRLEETLLLLHA 125
++A L TD+ V + +EE L +H+
Sbjct 68 NRIALLNTDYSNVMKEIEEKLFDIHS 93
> tpv:TP02_0299 hypothetical protein
Length=143
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 50/74 (67%), Gaps = 2/74 (2%)
Query 53 RQQIEEKMEALASFLTAE--GMPGLKGPLVDPEGFPRADIDVHAVLQARQQLACLKTDHR 110
R+ +E +MEAL S+L +E GL GPLVD + FPR DID++ V +AR ++ CLK D+
Sbjct 11 RKDVELEMEALMSYLNSEECKYVGLNGPLVDDDQFPRNDIDIYEVRKARGRIMCLKNDYE 70
Query 111 EVQQRLEETLLLLH 124
++ + +E+ L LH
Sbjct 71 KLTEEIEKLLHELH 84
> hsa:5715 PSMD9, MGC8644, Rpn4, p27; proteasome (prosome, macropain)
26S subunit, non-ATPase, 9; K06693 26S proteasome non-ATPase
regulatory subunit 9
Length=223
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 57/96 (59%), Gaps = 0/96 (0%)
Query 30 RRKMAASAGASSLAAQAEAVYRERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRAD 89
R+ +S + + + R +++IE +++A L ++ G+ PLVD EG+PR+D
Sbjct 7 RQSGGSSQAGVVTVSDVQELMRRKEEIEAQIKANYDVLESQKGIGMNEPLVDCEGYPRSD 66
Query 90 IDVHAVLQARQQLACLKTDHREVQQRLEETLLLLHA 125
+D++ V AR + CL+ DH+ V +++EE L LHA
Sbjct 67 VDLYQVRTARHNIICLQNDHKAVMKQVEEALHQLHA 102
> ath:AT5G57950 26S proteasome regulatory subunit, putative; K06693
26S proteasome non-ATPase regulatory subunit 9
Length=227
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 35/102 (34%), Positives = 59/102 (57%), Gaps = 7/102 (6%)
Query 39 ASSLAAQAEAVYRERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRADIDVHAVLQA 98
++L A+ A+ +R +E +M ++ L G PGL G L+D EGFPR DID+ V
Sbjct 3 GANLKAETMALMDKRTAMETEMNSIVERLCNPGGPGLSGNLIDSEGFPREDIDIPMVRTE 62
Query 99 RQQLACLKTDHREVQQRLEETLLLLH-------AANAKDKPP 133
R++LA L+++H E+ +++ + +LH A++ KD P
Sbjct 63 RRRLAELRSEHGEITEKINVNIQILHSVRPTSRASSTKDSGP 104
> pfa:PFC0785c proteasome regulatory protein, putative; K06693
26S proteasome non-ATPase regulatory subunit 9
Length=225
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 28/78 (35%), Positives = 50/78 (64%), Gaps = 2/78 (2%)
Query 51 RERQQIEEKMEALASFLT--AEGMPGLKGPLVDPEGFPRADIDVHAVLQARQQLACLKTD 108
++R+ IE +++ FL G+KG L+D EGFPR DID++++ AR ++ CLK D
Sbjct 11 KKREDIENELKEHMDFLERPENKNVGMKGNLIDSEGFPRNDIDIYSIRVARNKIICLKND 70
Query 109 HREVQQRLEETLLLLHAA 126
+ ++ ++LEE + +H+
Sbjct 71 YIDINKKLEEYIHKVHST 88
> mmu:67151 Psmd9, 1500011J20Rik, 2610202L11Rik, Bridge-1, P27;
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9;
K06693 26S proteasome non-ATPase regulatory subunit 9
Length=222
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 30/77 (38%), Positives = 51/77 (66%), Gaps = 0/77 (0%)
Query 49 VYRERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRADIDVHAVLQARQQLACLKTD 108
+ R +++IE +++A L ++ G+ PLVD EG+PRAD+D++ V AR + CL+ D
Sbjct 26 LMRRKEEIEAEIKANYDVLESQKGIGMNEPLVDCEGYPRADVDLYQVRTARHNIICLQND 85
Query 109 HREVQQRLEETLLLLHA 125
H+ + +++EE L LHA
Sbjct 86 HKALMKQVEEALHQLHA 102
> xla:379096 MGC53232; similar to proteasome (prosome, macropain)
26S subunit, non-ATPase, 9; K06693 26S proteasome non-ATPase
regulatory subunit 9
Length=213
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 46/74 (62%), Gaps = 0/74 (0%)
Query 52 ERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRADIDVHAVLQARQQLACLKTDHRE 111
++ ++E +++AL L + G+ GPLVD EG+PR D+D+ V AR + CL+ DH+
Sbjct 23 KKDEMETQIKALYDLLQEQKGIGMDGPLVDREGYPRTDVDISQVRTARHNIICLQNDHKA 82
Query 112 VQQRLEETLLLLHA 125
+ + +E L LHA
Sbjct 83 IMKEIEVALHRLHA 96
> sce:YIL007C NAS2; Nas2p; K06693 26S proteasome non-ATPase regulatory
subunit 9
Length=220
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 42/79 (53%), Gaps = 2/79 (2%)
Query 53 RQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRADIDVHAVLQARQQLACLKTDHREV 112
+ IE ++EA S L +G+ G+ LV P+G+PR+D+DV V R+ + LK D +
Sbjct 38 KTDIETQLEAYFSVLEQQGI-GMDSALVTPDGYPRSDVDVLQVTMIRKNVNMLKNDLNHL 96
Query 113 QQRLEETLLLLHAANAKDK 131
QR LL H N K
Sbjct 97 LQR-SHVLLNQHFDNMNVK 114
> cel:C44B7.1 hypothetical protein
Length=197
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 38/65 (58%), Gaps = 1/65 (1%)
Query 44 AQAEAVYRERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRADIDVHAVLQARQQLA 103
++A+ + ++R +++ K++ L L + PL+D EG+P IDV+AV AR L
Sbjct 5 SKAKELLQQRDELDGKIKELMLVLETNN-STMDSPLLDAEGYPLNTIDVYAVRHARHDLI 63
Query 104 CLKTD 108
CL+ D
Sbjct 64 CLRND 68
> mmu:81000 Rad54l2, Arip4, D130058C05, G630026H09Rik, Srisnf2l;
RAD54 like 2 (S. cerevisiae) (EC:3.6.4.12); K10876 RAD54-like
protein 2 [EC:3.6.4.12]
Length=1467
Score = 31.6 bits (70), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 79 LVDPEGFPRADIDVHAVLQARQQLACLKTDHREVQQRLEETLLLLHAANAK 129
V+ E P+ +D+ + ++ QLACLK H ++ E LLL+ + K
Sbjct 909 FVEKEPAPQTSLDIKGIKESVLQLACLKYPHLITKEPFEHESLLLNRKDHK 959
> hsa:23132 RAD54L2, ARIP4, FLJ21396, FLJ22400, HSPC325, KIAA0809,
SRISNF2L; RAD54-like 2 (S. cerevisiae) (EC:3.6.4.12); K10876
RAD54-like protein 2 [EC:3.6.4.12]
Length=1467
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 79 LVDPEGFPRADIDVHAVLQARQQLACLKTDHREVQQRLEETLLLLHAANAK 129
V+ E P+ ++V + ++ QLACLK H ++ E LLL+ + K
Sbjct 909 FVEKEPAPQVSLNVKGIKESVLQLACLKYPHLITKEPFEHESLLLNRKDHK 959
> mmu:72236 Tsnaxip1, 1700016K08Rik, MGC118240, TXI1; translin-associated
factor X (Tsnax) interacting protein 1
Length=704
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 29/48 (60%), Gaps = 3/48 (6%)
Query 86 PRADIDVH--AVLQARQQLACLKTDHREVQQRLEETLLLLHAANAKDK 131
PR D+++ ++ ++QL LK D+ EVQ+ E LL LH + K++
Sbjct 303 PRRDLEMQEKTNMELQEQLESLKADYEEVQKE-HELLLQLHMSTLKER 349
> tgo:TGME49_089020 hypothetical protein
Length=451
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 36/69 (52%), Gaps = 4/69 (5%)
Query 37 AGASSLAAQAEAVYRE-RQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRADIDVHA- 94
A A LA QAE V E +++ E LA F E +G LVD EG R+ DV A
Sbjct 117 AKAEQLAKQAEGVQAECERRLSEHQRLLADFRVLEETH--RGLLVDMEGLSRSKEDVEAT 174
Query 95 VLQARQQLA 103
V + ++QLA
Sbjct 175 VAELQRQLA 183
> bbo:BBOV_II005840 18.m06484; hypothetical protein
Length=1146
Score = 30.0 bits (66), Expect = 2.3, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 32/79 (40%), Gaps = 5/79 (6%)
Query 14 LARFRSQSPRGCRLRPRRKMAASAGASSLAAQAEAVYRE-----RQQIEEKMEALASFLT 68
L R + +L PR + +LA + E Y R ++ K L +FL
Sbjct 390 LHRLTAPQTPSQQLFPRSPLDTCRAVQTLAEKVEEQYLSLSSFVRDKLVAKCRCLLAFLY 449
Query 69 AEGMPGLKGPLVDPEGFPR 87
G+P G L P G+PR
Sbjct 450 LFGVPDESGNLEIPGGWPR 468
> ath:AT1G02470 hypothetical protein
Length=221
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 38/84 (45%), Gaps = 5/84 (5%)
Query 30 RRKMAASAGASSLAAQAEAVYRERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRAD 89
R KM A AS A +Y +R+ + M L+S EG P L LV E F + +
Sbjct 82 RVKMVVDAPAS----VAYKLYADREMFPKWMPFLSSVEAMEGSPDLSRYLVKLESFGQ-N 136
Query 90 IDVHAVLQARQQLACLKTDHREVQ 113
I+ H + + Q + K R ++
Sbjct 137 IEYHFLAKNLQPIPDRKIHWRSIE 160
> hsa:10320 IKZF1, Hs.54452, IK1, IKAROS, LYF1, PRO0758, ZNFN1A1,
hIk-1; IKAROS family zinc finger 1 (Ikaros); K09220 IKAROS
family zinc finger protein
Length=519
Score = 29.3 bits (64), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 11/34 (32%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 50 YRERQQIEEKMEALASFLTAEGMPGLKGPLVDPE 83
Y++R +EE E ++L + G+PG P++ E
Sbjct 210 YKQRSSLEEHKERCHNYLESMGLPGTLYPVIKEE 243
> ath:AT5G14580 polyribonucleotide nucleotidyltransferase, putative;
K00962 polyribonucleotide nucleotidyltransferase [EC:2.7.7.8]
Length=991
Score = 28.5 bits (62), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 12/41 (29%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 28 RPRRKMAASAGASSLAAQAEAVYRERQQIEEKMEALASFLT 68
+P+RK A+ AG + ++ VY E + E + ++ S +T
Sbjct 751 KPKRKPASDAGKDPVMKESSTVYIENSSVGEIVASMPSIVT 791
Lambda K H
0.316 0.127 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40