bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2698_orf1
Length=199
Score E
Sequences producing significant alignments: (Bits) Value
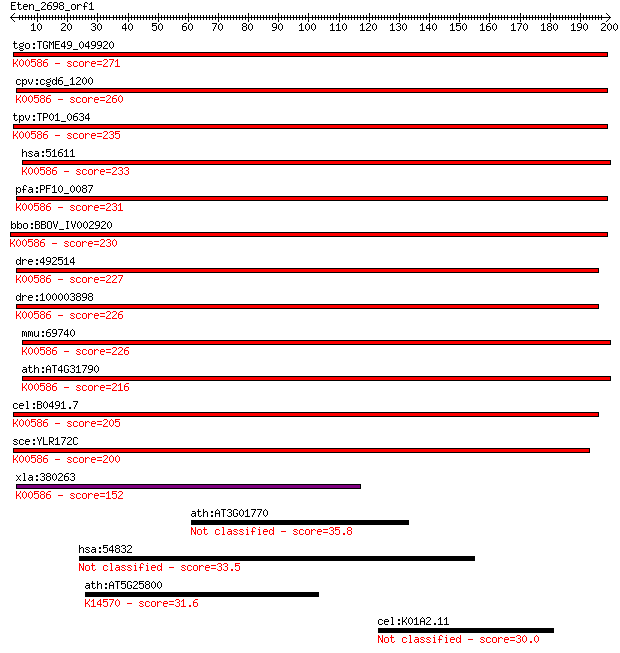
tgo:TGME49_049920 diphthine synthase, putative (EC:2.1.1.98); ... 271 1e-72
cpv:cgd6_1200 diphthine synthase; diphthamide biosynthesis met... 260 2e-69
tpv:TP01_0634 diphthine synthase; K00586 diphthine synthase [E... 235 8e-62
hsa:51611 DPH5, CGI-30, HSPC143, MGC61450, NPD015; DPH5 homolo... 233 5e-61
pfa:PF10_0087 diphthine synthase (EC:2.1.1.98); K00586 diphthi... 231 1e-60
bbo:BBOV_IV002920 21.m02908; diphthine synthase (EC:2.1.1.98);... 230 3e-60
dre:492514 dph5, zgc:101823; DPH5 homolog (S. cerevisiae) (EC:... 227 2e-59
dre:100003898 dph5; DPH5 homolog (S. cerevisiae); K00586 dipht... 226 3e-59
mmu:69740 Dph5, 2410012M04Rik, AU045680, C80186; DPH5 homolog ... 226 4e-59
ath:AT4G31790 diphthine synthase, putative (DPH5); K00586 diph... 216 3e-56
cel:B0491.7 hypothetical protein; K00586 diphthine synthase [E... 205 7e-53
sce:YLR172C DPH5; Dph5p (EC:2.1.1.98); K00586 diphthine syntha... 200 2e-51
xla:380263 dph5, MGC64452, cgi-30; DPH5 homolog; K00586 diphth... 152 5e-37
ath:AT3G01770 ATBET10 (Arabidopsis thaliana BROMODOMAIN AND EX... 35.8 0.11
hsa:54832 VPS13C, DKFZp686E0570, FLJ21361; vacuolar protein so... 33.5 0.55
ath:AT5G25800 exonuclease family protein; K14570 RNA exonuclea... 31.6 1.6
cel:K01A2.11 cbn-1; Calcium BiNding protein homolog family mem... 30.0 4.9
> tgo:TGME49_049920 diphthine synthase, putative (EC:2.1.1.98);
K00586 diphthine synthase [EC:2.1.1.98]
Length=275
Score = 271 bits (693), Expect = 1e-72, Method: Compositional matrix adjust.
Identities = 126/198 (63%), Positives = 155/198 (78%), Gaps = 1/198 (0%)
Query 2 TEAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQL 61
++ MLE A + VA LVVGDPFCATTH+DLYLRAR K VTV+V+HNASIM+A+ +CGLQL
Sbjct 66 SDEMLERALSSNVAFLVVGDPFCATTHADLYLRARKKNVTVRVVHNASIMNAIGSCGLQL 125
Query 62 YRFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEP 121
YRFGETVS+PFF+ W+P SFY KI+KN+E HTLCLLDIK KEQ+VEN+M+G I+EP
Sbjct 126 YRFGETVSIPFFEESWRPDSFYMKIKKNKEAGFHTLCLLDIKTKEQSVENMMRGRQIYEP 185
Query 122 PRFMTVNTAIRQLFEAAEMQGDEGVA-SILAFGLARVGADDQAIVSGPLQELLNADLGGP 180
PRFM+V A+RQL E + G + AFGLAR+GA Q I SG L+ELL+ D G P
Sbjct 186 PRFMSVEAAVRQLLEVEDKLGGKVCPRDAKAFGLARIGAPSQQITSGTLEELLSVDFGPP 245
Query 181 LHSLVLCAPELHEIEQKF 198
LHSLV+CAP LHE+E++F
Sbjct 246 LHSLVICAPTLHEMEREF 263
> cpv:cgd6_1200 diphthine synthase; diphthamide biosynthesis methyltransferase
; K00586 diphthine synthase [EC:2.1.1.98]
Length=274
Score = 260 bits (664), Expect = 2e-69, Method: Compositional matrix adjust.
Identities = 125/198 (63%), Positives = 151/198 (76%), Gaps = 3/198 (1%)
Query 3 EAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLY 62
+ ++E AK K + LVVGDPFCATTHSDL LRA +K V V+V HNASI+SA+ GLQ+Y
Sbjct 71 DEIIEEAKDKSIVLLVVGDPFCATTHSDLVLRAHEKDVKVEVRHNASIISAIGCTGLQVY 130
Query 63 RFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEPP 122
RFGETVS+PFFDG W+P SFY+KI+ N E +HTLCLLDIKVKEQT+EN+M+ IFEPP
Sbjct 131 RFGETVSIPFFDGSWQPSSFYDKIKANIERGLHTLCLLDIKVKEQTIENMMRNRPIFEPP 190
Query 123 RFMTVNTAIRQLFEAAEMQGDEGVAS--ILAFGLARVGADDQAIVSGPLQELLNADLGGP 180
RFMTVN AI QLF E + + V S LA G+AR+G+ DQ IVSG L EL + D G P
Sbjct 191 RFMTVNQAISQLF-ILEDKLKQNVISPNSLAIGVARIGSSDQKIVSGTLSELSDTDFGNP 249
Query 181 LHSLVLCAPELHEIEQKF 198
LHSLV+C P+LH IEQ+F
Sbjct 250 LHSLVICHPDLHLIEQRF 267
> tpv:TP01_0634 diphthine synthase; K00586 diphthine synthase
[EC:2.1.1.98]
Length=268
Score = 235 bits (599), Expect = 8e-62, Method: Compositional matrix adjust.
Identities = 106/197 (53%), Positives = 137/197 (69%), Gaps = 0/197 (0%)
Query 2 TEAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQL 61
+ +L AK K V L+ GDPF ATTH ++Y +A + G+ V +I+NASIM++V GLQL
Sbjct 66 NDTLLNEAKDKNVVLLIAGDPFSATTHVEIYYKAMNCGIDVNIINNASIMNSVGITGLQL 125
Query 62 YRFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEP 121
YRFGETVS+PFF+ WKP SFY+KI +N NN+HTLCLLDIKV+E++VEN+MK IFE
Sbjct 126 YRFGETVSIPFFEENWKPFSFYDKIMQNYNNNLHTLCLLDIKVRERSVENIMKNKLIFEE 185
Query 122 PRFMTVNTAIRQLFEAAEMQGDEGVASILAFGLARVGADDQAIVSGPLQELLNADLGGPL 181
PRFM++N AI QL + G+AR+ + DQ I SG LQ+LLN D G PL
Sbjct 186 PRFMSINKAIEQLLYVQSNNTTHSKIDFMVIGMARLCSKDQVIKSGKLQDLLNFDFGPPL 245
Query 182 HSLVLCAPELHEIEQKF 198
HSLV+C+P LH E+ F
Sbjct 246 HSLVICSPHLHHYEELF 262
> hsa:51611 DPH5, CGI-30, HSPC143, MGC61450, NPD015; DPH5 homolog
(S. cerevisiae) (EC:2.1.1.98); K00586 diphthine synthase
[EC:2.1.1.98]
Length=285
Score = 233 bits (593), Expect = 5e-61, Method: Compositional matrix adjust.
Identities = 112/200 (56%), Positives = 146/200 (73%), Gaps = 5/200 (2%)
Query 5 MLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLYRF 64
+L+ A VA LVVGDPF ATTHSDL LRA G+ +VIHNASIM+AV CGLQLY+F
Sbjct 68 ILKDADISDVAFLVVGDPFGATTHSDLVLRATKLGIPYRVIHNASIMNAVGCCGLQLYKF 127
Query 65 GETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEPPRF 124
GETVS+ F+ W+P SF++K++KNR+N MHTLCLLDIKVKEQ++ENL+KG I+EPPR+
Sbjct 128 GETVSIVFWTDTWRPESFFDKVKKNRQNGMHTLCLLDIKVKEQSLENLIKGRKIYEPPRY 187
Query 125 MTVNTAIRQLFEAAEMQ---GDEGVAS--ILAFGLARVGADDQAIVSGPLQELLNADLGG 179
M+VN A +QL E + Q G+E + L GLARVGADDQ I +G L+++ DLG
Sbjct 188 MSVNQAAQQLLEIVQNQRIRGEEPAVTEETLCVGLARVGADDQKIAAGTLRQMCTVDLGE 247
Query 180 PLHSLVLCAPELHEIEQKFV 199
PLHSL++ +H +E + +
Sbjct 248 PLHSLIITGGSIHPMEMEML 267
> pfa:PF10_0087 diphthine synthase (EC:2.1.1.98); K00586 diphthine
synthase [EC:2.1.1.98]
Length=274
Score = 231 bits (589), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 99/197 (50%), Positives = 141/197 (71%), Gaps = 1/197 (0%)
Query 3 EAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLY 62
+ +L+ AK K+V+ LVVGDP CATTH D+ LRA+ K + V++IHN SI+SA+ CG+QLY
Sbjct 67 DKILDEAKNKKVSFLVVGDPLCATTHHDIILRAKKKNIDVEIIHNTSIISAIGECGMQLY 126
Query 63 RFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEPP 122
FG+ VS+P+F+ +KP S+Y+KI N +NN HTLCLLDIKVKE+TVEN+M+ I+EPP
Sbjct 127 NFGQIVSIPYFEDNYKPTSYYDKIYINLKNNFHTLCLLDIKVKERTVENIMRNKKIYEPP 186
Query 123 RFMTVNTAIRQLFEAAEMQGDEGVA-SILAFGLARVGADDQAIVSGPLQELLNADLGGPL 181
RFMT+N +I QL + + + L + ++G D+Q I+SG L L + PL
Sbjct 187 RFMTINDSIEQLLYCEHIHKKNIITKNTLGIAIIQIGTDNQQIISGDLLTLKDISYNKPL 246
Query 182 HSLVLCAPELHEIEQKF 198
HSL++CAP LH+IE+++
Sbjct 247 HSLIICAPTLHDIEKEY 263
> bbo:BBOV_IV002920 21.m02908; diphthine synthase (EC:2.1.1.98);
K00586 diphthine synthase [EC:2.1.1.98]
Length=268
Score = 230 bits (586), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 107/198 (54%), Positives = 143/198 (72%), Gaps = 1/198 (0%)
Query 1 STEAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQ 60
S + +LE A+ K V LV GDP ATTH DL LRA + GV V+VIHNASI++A+ G+Q
Sbjct 66 SADKILEEARAKNVVLLVAGDPLSATTHCDLCLRAENAGVDVEVIHNASIINAIGRTGMQ 125
Query 61 LYRFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFE 120
LYRFGE VS+PFF+ W P SFY+KI KN E N+HTLCLLDIKV+E+++ENLM IFE
Sbjct 126 LYRFGEIVSIPFFETNWSPDSFYDKIVKNMEANLHTLCLLDIKVRERSIENLMNNRMIFE 185
Query 121 PPRFMTVNTAIRQLFEAAEMQGDEGVASILAFGLARVGADDQAIVSGPLQELLNADLGGP 180
PPR+M+VN AI Q+F + ++ A G+AR+G+ I +G L+EL + D G P
Sbjct 186 PPRYMSVNIAIDQIFRIDHTK-HRLPSNTRAIGVARLGSKTAKIAAGTLKELKDIDFGEP 244
Query 181 LHSLVLCAPELHEIEQKF 198
LHS+V+CAP+LH+IE+++
Sbjct 245 LHSMVICAPQLHDIEEEY 262
> dre:492514 dph5, zgc:101823; DPH5 homolog (S. cerevisiae) (EC:2.1.1.98);
K00586 diphthine synthase [EC:2.1.1.98]
Length=288
Score = 227 bits (579), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 112/198 (56%), Positives = 143/198 (72%), Gaps = 6/198 (3%)
Query 3 EAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLY 62
+ +L+ A VA LVVGDPF ATTHSDL LRA + G+ +VIHNAS+M+AV CGLQLY
Sbjct 66 DEILKGADVCDVAFLVVGDPFGATTHSDLVLRALNAGIQYRVIHNASVMNAVGCCGLQLY 125
Query 63 RFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEPP 122
FGETVS+ F+ W+P SFY+KI+KNR+ +HTLCLLDIKVKEQ++ENL++G I+EPP
Sbjct 126 NFGETVSIVFWTDTWRPESFYDKIKKNRDMGLHTLCLLDIKVKEQSMENLVRGRKIYEPP 185
Query 123 RFMTVNTAIRQLFEAAEMQGDEG-----VASILAFGLARVGADDQAIVSGPLQELLNADL 177
R+MTV A QL E + + D G + GLARVGA+DQ I SG L+EL + DL
Sbjct 186 RYMTVAQAAEQLLEILQNRRDRGEELAMTEDTVCVGLARVGAEDQTIRSGTLRELASCDL 245
Query 178 GGPLHSLVLCAPELHEIE 195
GGPLHSL++ + LH +E
Sbjct 246 GGPLHSLII-SGHLHPLE 262
> dre:100003898 dph5; DPH5 homolog (S. cerevisiae); K00586 diphthine
synthase [EC:2.1.1.98]
Length=288
Score = 226 bits (577), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 113/198 (57%), Positives = 142/198 (71%), Gaps = 6/198 (3%)
Query 3 EAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLY 62
+ +L+ A VA LVVGDPF ATTHSDL LRA + G+ +VIHNASIM+AV CGLQLY
Sbjct 66 DEILKGADVCDVAFLVVGDPFGATTHSDLVLRALNAGIQYRVIHNASIMNAVGCCGLQLY 125
Query 63 RFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEPP 122
FGETVS+ F+ W+P SFY+KI+KNR+ +HTLCLLDIKVKEQ++ENLM+G I+EPP
Sbjct 126 NFGETVSIVFWTDTWRPESFYDKIKKNRDMGLHTLCLLDIKVKEQSMENLMRGRKIYEPP 185
Query 123 RFMTVNTAIRQLFEAAEMQGDEG-----VASILAFGLARVGADDQAIVSGPLQELLNADL 177
R+MTV A QL E + + D G + GLARVGA+DQ I SG L+EL + DL
Sbjct 186 RYMTVAQAAEQLLEILQNRRDRGEELAMTEDTVCVGLARVGAEDQTIRSGTLRELASCDL 245
Query 178 GGPLHSLVLCAPELHEIE 195
GPLHSL++ + LH +E
Sbjct 246 EGPLHSLII-SGHLHPLE 262
> mmu:69740 Dph5, 2410012M04Rik, AU045680, C80186; DPH5 homolog
(S. cerevisiae) (EC:2.1.1.98); K00586 diphthine synthase [EC:2.1.1.98]
Length=281
Score = 226 bits (576), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 110/200 (55%), Positives = 143/200 (71%), Gaps = 5/200 (2%)
Query 5 MLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLYRF 64
+ + A VA LVVGDPF ATTHSDL LRA G+ +VIHNASIM+AV CGLQLYRF
Sbjct 68 IFKDADVSDVAFLVVGDPFGATTHSDLILRATKLGIPYQVIHNASIMNAVGCCGLQLYRF 127
Query 65 GETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEPPRF 124
GETVS+ F+ W+P SF++K+++NR N MHTLCLLDIKVKEQ++ENL++G I+EPPR+
Sbjct 128 GETVSIVFWTDTWRPESFFDKVKRNRANGMHTLCLLDIKVKEQSLENLIRGRKIYEPPRY 187
Query 125 MTVNTAIRQLFEAAE---MQGDEGVAS--ILAFGLARVGADDQAIVSGPLQELLNADLGG 179
M+VN A +QL E + +G+E + L GLARVGA+DQ I +G LQ++ LG
Sbjct 188 MSVNQAAQQLLEIVQNHRARGEEPAITEETLCVGLARVGAEDQKIAAGTLQQMCTVSLGE 247
Query 180 PLHSLVLCAPELHEIEQKFV 199
PLHSLV+ LH +E + +
Sbjct 248 PLHSLVITGGNLHPLEMEML 267
> ath:AT4G31790 diphthine synthase, putative (DPH5); K00586 diphthine
synthase [EC:2.1.1.98]
Length=277
Score = 216 bits (551), Expect = 3e-56, Method: Compositional matrix adjust.
Identities = 106/197 (53%), Positives = 139/197 (70%), Gaps = 3/197 (1%)
Query 5 MLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLYRF 64
M++ A VA LVVGDPF ATTHSDL +RA+ GV V+V+HNAS+M+AV CGLQLY +
Sbjct 73 MIDEAIDNDVAFLVVGDPFGATTHSDLVVRAKTLGVKVEVVHNASVMNAVGICGLQLYHY 132
Query 65 GETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNI-FEPPR 123
GETVS+PFF W+P SFYEKI+KNR +HTLCLLDI+VKE T E+L +G +EPPR
Sbjct 133 GETVSIPFFTETWRPDSFYEKIKKNRSLGLHTLCLLDIRVKEPTFESLCRGGKKQYEPPR 192
Query 124 FMTVNTAIRQLFEAAEMQGDEGVA-SILAFGLARVGADDQAIVSGPLQELLNADLGGPLH 182
+M+VNTAI QL E + GD G AR+G++DQ IV+G +++L + D G PLH
Sbjct 193 YMSVNTAIEQLLEVEQKHGDSVYGEDTQCVGFARLGSEDQTIVAGTMKQLESVDFGAPLH 252
Query 183 SLVLCAPELHEIEQKFV 199
LV+ E H +E++ +
Sbjct 253 CLVIVG-ETHPVEEEML 268
> cel:B0491.7 hypothetical protein; K00586 diphthine synthase
[EC:2.1.1.98]
Length=274
Score = 205 bits (522), Expect = 7e-53, Method: Compositional matrix adjust.
Identities = 103/199 (51%), Positives = 138/199 (69%), Gaps = 6/199 (3%)
Query 2 TEAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQL 61
++A+L A + VA LVVGDPF ATTH+DL LRA+ + + VKVIHNASIM+AV CGLQL
Sbjct 68 SDAILNGADKEDVALLVVGDPFGATTHADLVLRAKQQNIPVKVIHNASIMNAVGCCGLQL 127
Query 62 YRFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEP 121
Y FGETVS+ + W+P S+Y+KI NR+ MHTLCLLDIK KEQTVEN+M+G IFEP
Sbjct 128 YNFGETVSIVMWTDEWQPESYYDKIALNRKRGMHTLCLLDIKTKEQTVENMMRGRKIFEP 187
Query 122 PRFMTVNTAIRQL---FEAAEMQGDEGV--ASILAFGLARVGADDQAIVSGPLQELLNAD 176
R+ + A RQL +E + +G+E + + GLARVG D+Q IV ++++ +
Sbjct 188 ARYQKCSEAARQLLTIYERRKAKGEECAYDENTMVVGLARVGWDNQKIVYASMKDMSEME 247
Query 177 LGGPLHSLVLCAPELHEIE 195
+G PLHSL++ E H +E
Sbjct 248 MGEPLHSLII-PGETHPLE 265
> sce:YLR172C DPH5; Dph5p (EC:2.1.1.98); K00586 diphthine synthase
[EC:2.1.1.98]
Length=300
Score = 200 bits (509), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 95/192 (49%), Positives = 133/192 (69%), Gaps = 1/192 (0%)
Query 2 TEAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQL 61
++ +L A + VA LVVGDPF ATTH+DL LRA+ + + V++IHNAS+M+AV ACGLQL
Sbjct 66 SKQILNNADKEDVAFLVVGDPFGATTHTDLVLRAKREAIPVEIIHNASVMNAVGACGLQL 125
Query 62 YRFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEP 121
Y FG+TVS+ FF W+P S+Y+KI +NR+ +HTL LLDIKVKEQ++EN+ +G I+EP
Sbjct 126 YNFGQTVSMVFFTDNWRPDSWYDKIWENRKIGLHTLVLLDIKVKEQSIENMARGRLIYEP 185
Query 122 PRFMTVNTAIRQLFEAAEMQGDEGVA-SILAFGLARVGADDQAIVSGPLQELLNADLGGP 180
PR+M++ QL E E +G + A ++R+G+ Q+ SG + EL N D G P
Sbjct 186 PRYMSIAQCCEQLLEIEEKRGTKAYTPDTPAVAISRLGSSSQSFKSGTISELANYDSGEP 245
Query 181 LHSLVLCAPELH 192
LHSLV+ + H
Sbjct 246 LHSLVILGRQCH 257
> xla:380263 dph5, MGC64452, cgi-30; DPH5 homolog; K00586 diphthine
synthase [EC:2.1.1.98]
Length=241
Score = 152 bits (385), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 74/114 (64%), Positives = 86/114 (75%), Gaps = 0/114 (0%)
Query 3 EAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLY 62
+ +L A VA LVVGDPF ATTHSDL LRA G+ VIHNASIM+AV CGLQLY
Sbjct 66 DEILRDAAVSDVALLVVGDPFGATTHSDLVLRAAKAGIQHHVIHNASIMTAVGCCGLQLY 125
Query 63 RFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGN 116
FGETVS+ F+ WKP SFY+KI++NR + MHTLCLLDIKVKEQ++ENLM N
Sbjct 126 NFGETVSIVFWTDMWKPESFYDKIRRNRLSGMHTLCLLDIKVKEQSIENLMNFN 179
> ath:AT3G01770 ATBET10 (Arabidopsis thaliana BROMODOMAIN AND
EXTRATERMINAL DOMAIN PROTEIN 10); DNA binding
Length=620
Score = 35.8 bits (81), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 39/77 (50%), Gaps = 6/77 (7%)
Query 61 LYRFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVE----NLMKGN 116
+YRF +T+S FF+ WK + K+ +N+ TL DI + E + N +K N
Sbjct 212 VYRFADTLSK-FFEVRWKTIEKKSSGTKSEPSNLATLAHKDIAIPEPVAKKRKMNAVKRN 270
Query 117 NIFEP-PRFMTVNTAIR 132
++ EP R MT ++
Sbjct 271 SLLEPAKRVMTDEDRVK 287
> hsa:54832 VPS13C, DKFZp686E0570, FLJ21361; vacuolar protein
sorting 13 homolog C (S. cerevisiae)
Length=3628
Score = 33.5 bits (75), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 30/142 (21%), Positives = 61/142 (42%), Gaps = 23/142 (16%)
Query 24 CATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLYRFGETVSVPFFDGGWKPVSFY 83
C H + L + + + + S+M+AVA LQ++ + E +V W+P+
Sbjct 2289 CGLGHRTVPLLLAESKFSGNIKNWTSLMAAVADVTLQVHYYNEIHAV------WEPL--I 2340
Query 84 EKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEPPRF-----------MTVNTAIR 132
E+++ R+ N+ LD+K ++L+ G++ P+ +T++ +
Sbjct 2341 ERVEGKRQWNLR----LDVKKNPVQDKSLLPGDDFIPEPQMAIHISSGNTMNITISKSCL 2396
Query 133 QLFEAAEMQGDEGVASILAFGL 154
+F EG AS + L
Sbjct 2397 NVFNNLAKGFSEGTASTFDYSL 2418
> ath:AT5G25800 exonuclease family protein; K14570 RNA exonuclease
1 [EC:3.1.-.-]
Length=567
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 34/78 (43%), Gaps = 7/78 (8%)
Query 26 TTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLYRFGE-TVSVPFFDGGWKPVSFYE 84
T + L + R K + ++S+ AC L F E T +PF PVS+Y
Sbjct 129 TIDTILTCKGRKKKTVTSSVEPPPLVSSPEACNLMGKSFVELTKDIPF------PVSYYT 182
Query 85 KIQKNRENNMHTLCLLDI 102
QK E N +T L++
Sbjct 183 LSQKEMEQNGYTFEKLEL 200
> cel:K01A2.11 cbn-1; Calcium BiNding protein homolog family member
(cbn-1)
Length=679
Score = 30.0 bits (66), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 33/62 (53%), Gaps = 5/62 (8%)
Query 123 RFMTVNTAIRQLFEAAEMQGDEGV----ASILAFGLARVGADDQAIVSGPLQELLNADLG 178
R MT+ A RQ+ + A++ GD + A +AF +GA D A + G + E ++ +L
Sbjct 186 RGMTIRLA-RQIMKNADLNGDGHISVDEAQAIAFEQEGIGAGDVASMVGSVDENMDGELN 244
Query 179 GP 180
P
Sbjct 245 AP 246
Lambda K H
0.321 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5931269072
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40