bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
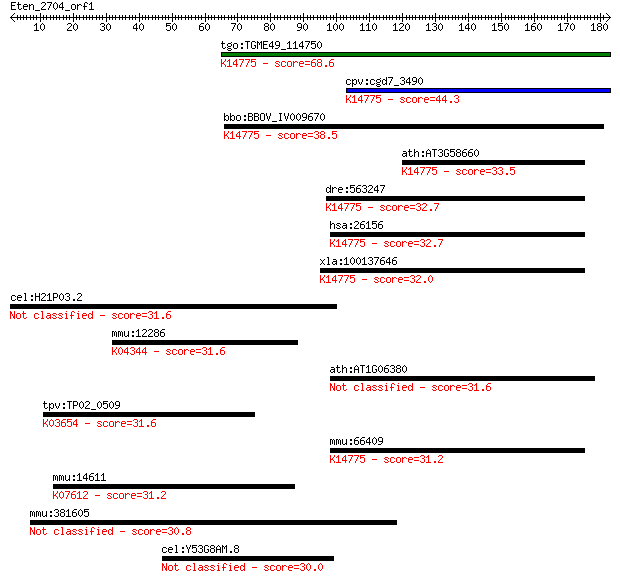
Query= Eten_2704_orf1
Length=182
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_114750 hypothetical protein ; K14775 ribosome bioge... 68.6 1e-11
cpv:cgd7_3490 hypothetical protein ; K14775 ribosome biogenesi... 44.3 2e-04
bbo:BBOV_IV009670 23.m06191; hypothetical protein; K14775 ribo... 38.5 0.013
ath:AT3G58660 60S ribosomal protein-related; K14775 ribosome b... 33.5 0.39
dre:563247 similar to ribosomal L1 domain containing 1; K14775... 32.7 0.67
hsa:26156 RSL1D1, CSIG, DKFZP564M182, MGC138433, MGC142259, PB... 32.7 0.71
xla:100137646 rsl1d1; ribosomal L1 domain containing 1; K14775... 32.0 1.1
cel:H21P03.2 hypothetical protein 31.6 1.4
mmu:12286 Cacna1a, APCA, Caca1a, Cacnl1a4, Cav2.1, Ccha1a, EA2... 31.6 1.5
ath:AT1G06380 ribosomal protein-related 31.6 1.7
tpv:TP02_0509 DNA helicase; K03654 ATP-dependent DNA helicase ... 31.6 1.7
mmu:66409 Rsl1d1, 2410005K20Rik, C76439, pBK1; ribosomal L1 do... 31.2 1.8
mmu:14611 Gja3, Cnx46, Cx43, Cx46, Gja-3; gap junction protein... 31.2 2.1
mmu:381605 Tbc1d2, A630005A06Rik, BC022630, Gm1037, PARIS-1, P... 30.8 2.8
cel:Y53G8AM.8 hypothetical protein 30.0 4.6
> tgo:TGME49_114750 hypothetical protein ; K14775 ribosome biogenesis
protein UTP30
Length=469
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 49/123 (39%), Positives = 60/123 (48%), Gaps = 24/123 (19%)
Query 65 AKSTPT-TAEKAKGCPQAPAASAPTVIKIGSGGVVDCQALEKAVKALCRHVQKLQEKEEV 123
A+++P T EK GCP +PA L+KA+ L +V K +
Sbjct 92 ARASPAETEEKIPGCPVSPAL------------------LKKALAGLTAYVMKKASERGT 133
Query 124 PDLLAMNSGPTVSLMFSTQNIPVKQRIYPHLIELPHSPLASNS----EVCLIVRDPQRKW 179
DLL TVSLM + +NIP R P I LPH PL S E CL V+DPQRKW
Sbjct 134 QDLLEETGAATVSLMLALKNIPQAFRKQPVQIVLPH-PLFDVSKGEGECCLFVKDPQRKW 192
Query 180 KDL 182
KDL
Sbjct 193 KDL 195
> cpv:cgd7_3490 hypothetical protein ; K14775 ribosome biogenesis
protein UTP30
Length=250
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/84 (40%), Positives = 51/84 (60%), Gaps = 10/84 (11%)
Query 103 LEKAVKALCRHVQKLQEKEEVPDLLAMNSGPTVSLMFSTQNIP--VKQRIYPHL-IELPH 159
LE AV AL + + K+++ E DLL + ++L+ S N P VK + H+ I++P+
Sbjct 16 LEDAVNAL-KELVKIKKCPE--DLLFNKNSQFITLIISLSNTPEFVKSK---HVRIKIPN 69
Query 160 SPLAS-NSEVCLIVRDPQRKWKDL 182
S +EVCL V+DPQRKWK+L
Sbjct 70 SLYKHPGNEVCLFVKDPQRKWKEL 93
> bbo:BBOV_IV009670 23.m06191; hypothetical protein; K14775 ribosome
biogenesis protein UTP30
Length=311
Score = 38.5 bits (88), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 37/123 (30%), Positives = 58/123 (47%), Gaps = 14/123 (11%)
Query 66 KSTPTTAEKAKGCPQAPAASAPTVI--KIGSGGV------VDCQALEKAVKALCRHVQKL 117
K T KA G P AS P I K+G + V ++++KAV AL + +
Sbjct 20 KKTRKEISKASGKP----ASKPVSIIPKVGVSALDNEHITVTEESIKKAVSALKKRAEAE 75
Query 118 QEKEEVPDLLAMNSGPTVSLMFSTQNIPVKQRIYPHLIELPHSPLASNSEVCLIVRDPQR 177
+E + DLL S V L + + I P I+L + P+ ++C+ V+DPQ+
Sbjct 76 RESRTL-DLLEDPSRCYVHLQIFLHKVFPETHIKPLKIQLQY-PIYRGKDICIFVKDPQK 133
Query 178 KWK 180
+WK
Sbjct 134 EWK 136
> ath:AT3G58660 60S ribosomal protein-related; K14775 ribosome
biogenesis protein UTP30
Length=446
Score = 33.5 bits (75), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 29/58 (50%), Gaps = 5/58 (8%)
Query 120 KEEVPDLLAMNSGPTVSLMFSTQNIPVKQRIYPHLIELPH---SPLASNSEVCLIVRD 174
K E P LL + V L + + IP K R + I LPH +P + E+CLI+ D
Sbjct 45 KTEKPQLLEEDE--LVYLFVTLKKIPQKTRTNAYRIPLPHPLINPTVDSPEICLIIDD 100
> dre:563247 similar to ribosomal L1 domain containing 1; K14775
ribosome biogenesis protein UTP30
Length=373
Score = 32.7 bits (73), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 37/78 (47%), Gaps = 6/78 (7%)
Query 97 VVDCQALEKAVKALCRHVQKLQEKEEVPDLLAMNSGPTVSLMFSTQNIPVKQRIYPHLIE 156
+D ++KA +AL ++ ++ L N G +SL+F+ IP K++ I
Sbjct 20 TIDQSQVKKAAQALQAFIKNSTSSQK----LFQNDGQPISLLFTLWKIPKKEQTIR--IP 73
Query 157 LPHSPLASNSEVCLIVRD 174
L H + +VCL RD
Sbjct 74 LSHGLRTESGDVCLFTRD 91
> hsa:26156 RSL1D1, CSIG, DKFZP564M182, MGC138433, MGC142259,
PBK1; ribosomal L1 domain containing 1; K14775 ribosome biogenesis
protein UTP30
Length=490
Score = 32.7 bits (73), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 38/78 (48%), Gaps = 8/78 (10%)
Query 98 VDCQALEKAVKALCRHVQKLQEKEEVPDLLAMNSGPTVSLMFSTQNIPVKQ-RIYPHLIE 156
+D + + KAV AL H + + L +N ++ LM IP K+ R+ +
Sbjct 31 LDKEQVRKAVDALLTHCKSRKNNYG----LLLNENESLFLMVVLWKIPSKELRVR---LT 83
Query 157 LPHSPLASNSEVCLIVRD 174
LPHS + + ++CL +D
Sbjct 84 LPHSIRSDSEDICLFTKD 101
> xla:100137646 rsl1d1; ribosomal L1 domain containing 1; K14775
ribosome biogenesis protein UTP30
Length=318
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 40/80 (50%), Gaps = 6/80 (7%)
Query 95 GGVVDCQALEKAVKALCRHVQKLQEKEEVPDLLAMNSGPTVSLMFSTQNIPVKQRIYPHL 154
G +D ++KAV+AL + Q+ + + L +N +S+M + IP +++
Sbjct 6 GHELDSAQVKKAVQALLAY----QKNKGNANSLLLNEHDRISMMLTVWRIPPREQAIR-- 59
Query 155 IELPHSPLASNSEVCLIVRD 174
I LPH+ +VCL RD
Sbjct 60 IPLPHAIRPEVCDVCLFTRD 79
> cel:H21P03.2 hypothetical protein
Length=397
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 45/99 (45%), Gaps = 11/99 (11%)
Query 1 TGRCRLFGCFISNEVTMIKVKKRKNGIAAAAKPLSVDTPDGKHKAKRSSSAPQSLSSPSQ 60
T +CR N ++KK+ + +A K +VD+ K K+ SA SL S+
Sbjct 40 TVQCR------ENHAVAEELKKQISELAHIRKSETVDS-----KIKKLESAVTSLREQSK 88
Query 61 PRKKAKSTPTTAEKAKGCPQAPAASAPTVIKIGSGGVVD 99
P KKA +T + + + T+++I G V+D
Sbjct 89 PLKKALNTALYSTDTELLELVGQTTRETLLEINGGAVID 127
> mmu:12286 Cacna1a, APCA, Caca1a, Cacnl1a4, Cav2.1, Ccha1a, EA2,
FHM, HPCA, MHP, MHP1, SCA6, alpha1A, la, leaner, nmf352,
rkr, rocker, tg, tottering; calcium channel, voltage-dependent,
P/Q type, alpha 1A subunit; K04344 voltage-dependent calcium
channel P/Q type alpha-1A
Length=2368
Score = 31.6 bits (70), Expect = 1.5, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 28/57 (49%), Gaps = 2/57 (3%)
Query 32 KPLSVDTPDGKHKAKRSSSAPQSLSSPSQPRKKAKSTPTTAEKAKGCPQ-APAASAP 87
+PL VD + ++ S AP++L ++PR+ A+ P G P+ AP P
Sbjct 822 RPLVVDPQENRNNNTNKSRAPEALRPTARPRESARD-PDARRAWPGSPERAPGREGP 877
> ath:AT1G06380 ribosomal protein-related
Length=254
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 40/85 (47%), Gaps = 9/85 (10%)
Query 98 VDCQALEKAVKALCRHVQKLQEKEEVPDLLAMNSGPTVSLMFSTQNIPVKQRIYPHLIEL 157
VD Q + +AVK+L + + E L N G V L+ + + IP R P +I L
Sbjct 18 VDPQNVNRAVKSLLKWWDSKSKTENSESL--ENDG-FVYLIVTLKRIPQLDRTNPLMIPL 74
Query 158 PHSPLASNS-----EVCLIVRDPQR 177
PH PL E+CLI+ D +
Sbjct 75 PH-PLIDLVAEDPPELCLIIDDKHK 98
> tpv:TP02_0509 DNA helicase; K03654 ATP-dependent DNA helicase
RecQ [EC:3.6.4.12]
Length=998
Score = 31.6 bits (70), Expect = 1.7, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 36/64 (56%), Gaps = 9/64 (14%)
Query 11 ISNEVTMIKVKKRKNGIAAAAKPLSVDTPDGKHKAKRSSSAPQSLSSPSQPRKKAKSTPT 70
++ + T IK + + +G A + + T + + K++R+SSA +P +PRKK+ TP
Sbjct 908 LNTDETPIKPRGKSDGTAKSRR----KTGETQPKSRRTSSA-----TPRKPRKKSDDTPV 958
Query 71 TAEK 74
+ K
Sbjct 959 KSRK 962
> mmu:66409 Rsl1d1, 2410005K20Rik, C76439, pBK1; ribosomal L1
domain containing 1; K14775 ribosome biogenesis protein UTP30
Length=452
Score = 31.2 bits (69), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 39/78 (50%), Gaps = 8/78 (10%)
Query 98 VDCQALEKAVKALCRHVQKLQEKEEVPDLLAMNSGPTVSLMFSTQNIPVKQ-RIYPHLIE 156
+D + + KAV+ + + + E L ++ + LM IP K+ R+ +
Sbjct 31 LDREQIRKAVEVISNRSKSKKNNNE----LLLSGSENLFLMVILWKIPEKELRVK---VP 83
Query 157 LPHSPLASNSEVCLIVRD 174
LPHS L+ +S+VCL +D
Sbjct 84 LPHSILSESSDVCLFTKD 101
> mmu:14611 Gja3, Cnx46, Cx43, Cx46, Gja-3; gap junction protein,
alpha 3; K07612 gap junction protein, alpha 3
Length=417
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 33/78 (42%), Gaps = 8/78 (10%)
Query 14 EVTMIKVKKRKNGIAAAAKPLSVDTPDGKHKA-----KRSSSAPQSLSSPSQPRKKAKST 68
E+ + KK K G+ P D + +HK +SS P S+S P +
Sbjct 227 EIYHLGWKKLKQGVTNHFNP---DASEARHKPLDPLPTATSSGPPSVSIGFPPYYTHPAC 283
Query 69 PTTAEKAKGCPQAPAASA 86
PT KA G P AP + A
Sbjct 284 PTVQAKAIGFPGAPLSPA 301
> mmu:381605 Tbc1d2, A630005A06Rik, BC022630, Gm1037, PARIS-1,
PARIS1; TBC1 domain family, member 2
Length=922
Score = 30.8 bits (68), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 34/119 (28%), Positives = 49/119 (41%), Gaps = 19/119 (15%)
Query 7 FGCFISNEVTMIKVKKRKNGIAAAAKPLSVDTPDGKHKAKRSSSAPQSLSSPSQPRKKAK 66
G I N TM ++ K AAA PL D+P G + S S+S PS P K+ +
Sbjct 214 LGTEIQN--TMHNIRGNKQAQAAAHGPLVEDSPQG---GEPQSGEQPSISDPSLPEKEPE 268
Query 67 STPTTAEKAKGCPQAPAASAPT--------VIKIGSGGVVDCQALEKAVKALCRHVQKL 117
+ AK P++ S PT S G+ + ++ V AL + V L
Sbjct 269 ------DPAKSAPRSSVPSGPTQKPKRQSNTFPFFSDGLARSRTAQEKVAALEQQVLML 321
> cel:Y53G8AM.8 hypothetical protein
Length=839
Score = 30.0 bits (66), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Query 47 RSSSAPQSLSSPSQPRKKAKSTPTTAEKAKGCPQAPAASAPTVIKIGSGGVV 98
R +S +S PS RKK STPT + PQ P+ SA TV + +G +
Sbjct 16 RGNSGARSHKEPSAKRKKPTSTPTV--RCGTVPQEPSTSAATVTQKPAGSTM 65
Lambda K H
0.314 0.128 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4912245712
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40