bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2711_orf1
Length=175
Score E
Sequences producing significant alignments: (Bits) Value
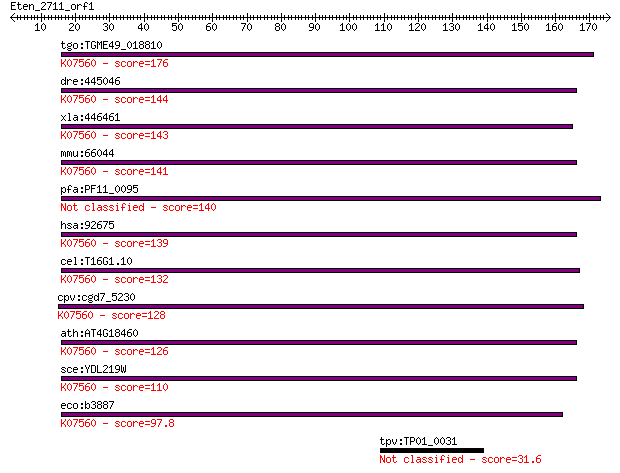
tgo:TGME49_018810 histidyl tRNA synthetase 2 ; K07560 D-tyrosy... 176 3e-44
dre:445046 hars2, zgc:92657; histidyl-tRNA synthetase 2, mitoc... 144 1e-34
xla:446461 dtd1, MGC78931, hars2; D-tyrosyl-tRNA deacylase 1 h... 143 3e-34
mmu:66044 Dtd1, 0610006H08Rik, Hars2; D-tyrosyl-tRNA deacylase... 141 1e-33
pfa:PF11_0095 D-tyrosyl-tRNA(Tyr) deacylase, putative 140 2e-33
hsa:92675 DTD1, C20orf88, DUEB, HARS2, MGC119131, MGC41905, bA... 139 5e-33
cel:T16G1.10 pqn-68; Prion-like-(Q/N-rich)-domain-bearing prot... 132 4e-31
cpv:cgd7_5230 possible D-Tyr-tRNatyr deacylase ; K07560 D-tyro... 128 8e-30
ath:AT4G18460 D-Tyr-tRNA(Tyr) deacylase family protein; K07560... 126 4e-29
sce:YDL219W DTD1; D-tyrosyl-tRNA(Tyr) deacylase (EC:3.1.-.-); ... 110 3e-24
eco:b3887 dtd, ECK3880, JW3858, yihZ; D-tyr-tRNA(Tyr) deacylas... 97.8 2e-20
tpv:TP01_0031 glutamyl-tRNA amidotransferase subunit B 31.6 1.6
> tgo:TGME49_018810 histidyl tRNA synthetase 2 ; K07560 D-tyrosyl-tRNA(Tyr)
deacylase [EC:3.1.-.-]
Length=223
Score = 176 bits (447), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 81/157 (51%), Positives = 107/157 (68%), Gaps = 2/157 (1%)
Query 16 MKLILQRVEGAAVKVAETGEEVARIGKGLICLLGIGKHDHKADVDYGVKKCLGTRLWED- 74
M+++LQRVE A V+V ETGE +IG+G++CLLGI D D DY ++KCL +RLW+D
Sbjct 1 MRMVLQRVESACVQVVETGELAGKIGRGIVCLLGISGEDKWEDADYCIRKCLKSRLWDDV 60
Query 75 -SSGKPWQKSVVDMGYELLLVSNFTLQAQTKKGFQPDFSAAMPPDKARPVYESFLENLRT 133
K W VVD YE+L+VS FTL KKG +PDF AAM PD+AR ++E + +R
Sbjct 61 KDPSKSWASCVVDRDYEVLVVSQFTLMGHLKKGNKPDFHAAMSPDQARSLFEKVVAEMRR 120
Query 134 GYKPSKVQCGQFQTRMRVEIHNDGPVTLTIDTQTLQL 170
YKP K+Q G+FQ MRVE+ NDGPVT+ +D+ QL
Sbjct 121 QYKPEKIQTGKFQNYMRVELVNDGPVTILVDSTQAQL 157
> dre:445046 hars2, zgc:92657; histidyl-tRNA synthetase 2, mitochondrial
(putative) (EC:6.1.1.21); K07560 D-tyrosyl-tRNA(Tyr)
deacylase [EC:3.1.-.-]
Length=207
Score = 144 bits (364), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 73/150 (48%), Positives = 101/150 (67%), Gaps = 3/150 (2%)
Query 16 MKLILQRVEGAAVKVAETGEEVARIGKGLICLLGIGKHDHKADVDYGVKKCLGTRLWEDS 75
MK I+QRV A+V V E E+++ IG+GL LLGI D + DVDY V+K L R++ED
Sbjct 1 MKAIIQRVTRASVTVGE--EQISSIGRGLCVLLGISAEDTQKDVDYMVRKILNLRVFEDE 58
Query 76 SGKPWQKSVVDMGYELLLVSNFTLQAQTKKGFQPDFSAAMPPDKARPVYESFLENLRTGY 135
+G+ W +SV+D E+L VS FTLQ KG +PD+ AAMP + A+P Y + LE LR Y
Sbjct 59 NGRAWSRSVMDGELEVLCVSQFTLQC-LLKGNKPDYHAAMPAELAQPFYNNMLEQLRETY 117
Query 136 KPSKVQCGQFQTRMRVEIHNDGPVTLTIDT 165
KP ++ GQF +M+V I NDGPVT+ +++
Sbjct 118 KPELIKDGQFGAKMQVLIQNDGPVTIQLES 147
> xla:446461 dtd1, MGC78931, hars2; D-tyrosyl-tRNA deacylase 1
homolog (EC:6.1.1.21); K07560 D-tyrosyl-tRNA(Tyr) deacylase
[EC:3.1.-.-]
Length=207
Score = 143 bits (360), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 68/149 (45%), Positives = 101/149 (67%), Gaps = 3/149 (2%)
Query 16 MKLILQRVEGAAVKVAETGEEVARIGKGLICLLGIGKHDHKADVDYGVKKCLGTRLWEDS 75
M+ ++QRV A+V V + E+++ IG+G+ LLGI D + D++Y V+K L RL+ D
Sbjct 1 MRAVIQRVTKASVTVGD--EQISSIGRGICVLLGISVEDTQKDIEYMVRKILNLRLFTDE 58
Query 76 SGKPWQKSVVDMGYELLLVSNFTLQAQTKKGFQPDFSAAMPPDKARPVYESFLENLRTGY 135
SGKPW KSV+D YE+L VS FTLQ KG +PD+ AMP ++A P Y +FL+++R Y
Sbjct 59 SGKPWCKSVMDKQYEVLCVSQFTLQC-VLKGNKPDYHMAMPSEQAEPFYNNFLQHMRKAY 117
Query 136 KPSKVQCGQFQTRMRVEIHNDGPVTLTID 164
KP ++ G+F M++ I NDGPVT+ ++
Sbjct 118 KPELIKDGKFGAYMQLNIQNDGPVTIELE 146
> mmu:66044 Dtd1, 0610006H08Rik, Hars2; D-tyrosyl-tRNA deacylase
1 homolog (S. cerevisiae) (EC:6.1.1.21); K07560 D-tyrosyl-tRNA(Tyr)
deacylase [EC:3.1.-.-]
Length=209
Score = 141 bits (356), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 70/150 (46%), Positives = 100/150 (66%), Gaps = 3/150 (2%)
Query 16 MKLILQRVEGAAVKVAETGEEVARIGKGLICLLGIGKHDHKADVDYGVKKCLGTRLWEDS 75
MK ++QRV A+V V GE+++ IG+G+ LLGI D + ++++ V+K L R++ED
Sbjct 1 MKAVVQRVTRASVTVG--GEQISAIGRGICVLLGISMEDSQKELEHMVRKILNLRVFEDE 58
Query 76 SGKPWQKSVVDMGYELLLVSNFTLQAQTKKGFQPDFSAAMPPDKARPVYESFLENLRTGY 135
SGK W KSV+D YE+L VS FTLQ KG +PDF AMP ++A Y SFLE LR Y
Sbjct 59 SGKHWSKSVMDKEYEVLCVSQFTLQC-VLKGNKPDFHLAMPTEQAESFYNSFLEQLRKSY 117
Query 136 KPSKVQCGQFQTRMRVEIHNDGPVTLTIDT 165
+P ++ G+F M+V I NDGPVT+ +++
Sbjct 118 RPELIRDGKFGAYMQVHIQNDGPVTIELES 147
> pfa:PF11_0095 D-tyrosyl-tRNA(Tyr) deacylase, putative
Length=164
Score = 140 bits (354), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 67/165 (40%), Positives = 99/165 (60%), Gaps = 9/165 (5%)
Query 16 MKLILQRVEGAAVKVAETG--------EEVARIGKGLICLLGIGKHDHKADVDYGVKKCL 67
M++++QRV+GA + V + E ++ I GLIC LGI K+D D Y ++KCL
Sbjct 1 MRVVIQRVKGAILSVRKENIGENEKELEIISEIKNGLICFLGIHKNDTWEDALYIIRKCL 60
Query 68 GTRLWEDSSGKPWQKSVVDMGYELLLVSNFTLQAQTKKGFQPDFSAAMPPDKARPVYESF 127
RLW ++ K W K+V D+ YELL+VS FTL TKKG +PDF A P++A Y
Sbjct 61 NLRLW-NNDNKTWDKNVKDLNYELLIVSQFTLFGNTKKGNKPDFHLAKEPNEALIFYNKI 119
Query 128 LENLRTGYKPSKVQCGQFQTRMRVEIHNDGPVTLTIDTQTLQLNR 172
++ + Y K++ G+F M +++ NDGPVT+ IDT + LN+
Sbjct 120 IDEFKKQYNDDKIKIGKFGNYMNIDVTNDGPVTIYIDTHDINLNK 164
> hsa:92675 DTD1, C20orf88, DUEB, HARS2, MGC119131, MGC41905,
bA379J5.3, bA555E18.1, pqn-68; D-tyrosyl-tRNA deacylase 1 homolog
(S. cerevisiae) (EC:6.1.1.21); K07560 D-tyrosyl-tRNA(Tyr)
deacylase [EC:3.1.-.-]
Length=209
Score = 139 bits (350), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 70/150 (46%), Positives = 100/150 (66%), Gaps = 3/150 (2%)
Query 16 MKLILQRVEGAAVKVAETGEEVARIGKGLICLLGIGKHDHKADVDYGVKKCLGTRLWEDS 75
MK ++QRV A+V V GE+++ IG+G+ LLGI D + ++++ V+K L R++ED
Sbjct 1 MKAVVQRVTRASVTVG--GEQISAIGRGICVLLGISLEDTQKELEHMVRKILNLRVFEDE 58
Query 76 SGKPWQKSVVDMGYELLLVSNFTLQAQTKKGFQPDFSAAMPPDKARPVYESFLENLRTGY 135
SGK W KSV+D YE+L VS FTLQ KG +PDF AMP ++A Y SFLE LR Y
Sbjct 59 SGKHWSKSVMDKQYEILCVSQFTLQC-VLKGNKPDFHLAMPTEQAEGFYNSFLEQLRKTY 117
Query 136 KPSKVQCGQFQTRMRVEIHNDGPVTLTIDT 165
+P ++ G+F M+V I NDGPVT+ +++
Sbjct 118 RPELIKDGKFGAYMQVHIQNDGPVTIELES 147
> cel:T16G1.10 pqn-68; Prion-like-(Q/N-rich)-domain-bearing protein
family member (pqn-68); K07560 D-tyrosyl-tRNA(Tyr) deacylase
[EC:3.1.-.-]
Length=150
Score = 132 bits (333), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 70/151 (46%), Positives = 96/151 (63%), Gaps = 3/151 (1%)
Query 16 MKLILQRVEGAAVKVAETGEEVARIGKGLICLLGIGKHDHKADVDYGVKKCLGTRLWEDS 75
MK++LQRV AAV V + E V IG+GL L+G+ + D + D+ Y ++K L R++ S
Sbjct 1 MKVVLQRVTRAAVTVGD--EVVGSIGRGLCVLVGVHRDDTEEDMKYIIRKILNLRVFPAS 58
Query 76 SGKPWQKSVVDMGYELLLVSNFTLQAQTKKGFQPDFSAAMPPDKARPVYESFLENLRTGY 135
KPW KSV+D+ E+L VS FTL Q KG + DF AM P +A Y SFLE ++ Y
Sbjct 59 EQKPWDKSVMDLDLEVLSVSQFTLYGQF-KGNKLDFHTAMAPTEASKFYASFLEAMKKAY 117
Query 136 KPSKVQCGQFQTRMRVEIHNDGPVTLTIDTQ 166
K K+Q G+F M V+I NDGPVT+T D++
Sbjct 118 KADKIQDGKFAAMMSVDIVNDGPVTVTFDSK 148
> cpv:cgd7_5230 possible D-Tyr-tRNatyr deacylase ; K07560 D-tyrosyl-tRNA(Tyr)
deacylase [EC:3.1.-.-]
Length=178
Score = 128 bits (322), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 66/157 (42%), Positives = 96/157 (61%), Gaps = 6/157 (3%)
Query 15 KMKLILQRVEGAAVKVAETGEEVARIGKGLICLLGIGKHDHKADVDYGVKKCLGTRLWED 74
+MK++LQ+V+GA+VKV G+ V+ IG GL+ L+GI D ++ DY V+KCL RLW D
Sbjct 12 RMKVVLQKVKGASVKV--DGQIVSSIGPGLLLLVGIRTDDVDSNSDYLVRKCLSIRLWPD 69
Query 75 ----SSGKPWQKSVVDMGYELLLVSNFTLQAQTKKGFQPDFSAAMPPDKARPVYESFLEN 130
+S KPW+ SV D E+L+VS FTL K G +PDF AM A ++ + +E
Sbjct 70 ESDPTSSKPWKLSVKDKDLEVLVVSQFTLFGNVKNGSKPDFHNAMSGKDALVIFNNMVEK 129
Query 131 LRTGYKPSKVQCGQFQTRMRVEIHNDGPVTLTIDTQT 167
+ + P K++ G F M V + NDGPVTL ++ +
Sbjct 130 FKKSHDPEKIKTGCFGEEMEVSLVNDGPVTLILENNS 166
> ath:AT4G18460 D-Tyr-tRNA(Tyr) deacylase family protein; K07560
D-tyrosyl-tRNA(Tyr) deacylase [EC:3.1.-.-]
Length=153
Score = 126 bits (316), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 65/151 (43%), Positives = 95/151 (62%), Gaps = 4/151 (2%)
Query 16 MKLILQRVEGAAVKVAETGEEVARIGKGLICLLGIGKHDHKADVDYGVKKCLGTRLW-ED 74
M+ ++QRV ++V V G V+ IG GL+ L+GI + D ++D DY +K L RL+ +
Sbjct 1 MRAVIQRVSSSSVTV--DGRIVSEIGPGLLVLIGIHESDTESDADYICRKVLNMRLFSNE 58
Query 75 SSGKPWQKSVVDMGYELLLVSNFTLQAQTKKGFQPDFSAAMPPDKARPVYESFLENLRTG 134
++GK W ++V+ Y +LLVS FTL K G +PDF AMPPDKA+P Y S +E +
Sbjct 59 TTGKGWDQNVMQRNYGVLLVSQFTLYGFLK-GNKPDFHVAMPPDKAKPFYASLVEKFQKA 117
Query 135 YKPSKVQCGQFQTRMRVEIHNDGPVTLTIDT 165
Y P V+ G F M+V + NDGPVT+ +D+
Sbjct 118 YNPDAVKDGVFGAMMQVNLVNDGPVTMQLDS 148
> sce:YDL219W DTD1; D-tyrosyl-tRNA(Tyr) deacylase (EC:3.1.-.-);
K07560 D-tyrosyl-tRNA(Tyr) deacylase [EC:3.1.-.-]
Length=150
Score = 110 bits (274), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 59/150 (39%), Positives = 90/150 (60%), Gaps = 2/150 (1%)
Query 16 MKLILQRVEGAAVKVAETGEEVARIGKGLICLLGIGKHDHKADVDYGVKKCLGTRLWEDS 75
MK++LQ+V A+V V + ++ I G + L+GI D A++D KK L R++ED
Sbjct 1 MKIVLQKVSQASVVV--DSKVISSIKHGYMLLVGISIDDSMAEIDKLSKKVLSLRIFEDE 58
Query 76 SGKPWQKSVVDMGYELLLVSNFTLQAQTKKGFQPDFSAAMPPDKARPVYESFLENLRTGY 135
S W+K++ + E+L VS FTL A+TKKG +PDF A A+ +YE FL+ LR+
Sbjct 59 SRNLWKKNIKEANGEILSVSQFTLMAKTKKGTKPDFHLAQKGHIAKELYEEFLKLLRSDL 118
Query 136 KPSKVQCGQFQTRMRVEIHNDGPVTLTIDT 165
KV+ G+F M + N+GPVT+ +D+
Sbjct 119 GEEKVKDGEFGAMMSCSLTNEGPVTIILDS 148
> eco:b3887 dtd, ECK3880, JW3858, yihZ; D-tyr-tRNA(Tyr) deacylase
(EC:3.1.-.-); K07560 D-tyrosyl-tRNA(Tyr) deacylase [EC:3.1.-.-]
Length=145
Score = 97.8 bits (242), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 56/146 (38%), Positives = 82/146 (56%), Gaps = 5/146 (3%)
Query 16 MKLILQRVEGAAVKVAETGEEVARIGKGLICLLGIGKHDHKADVDYGVKKCLGTRLWEDS 75
M ++QRV A+V V GE IG GL+ LLG+ K D + + ++ LG R++ D+
Sbjct 1 MIALIQRVTRASVTV--EGEVTGEIGAGLLVLLGVEKDDDEQKANRLCERVLGYRIFSDA 58
Query 76 SGKPWQKSVVDMGYELLLVSNFTLQAQTKKGFQPDFSAAMPPDKARPVYESFLENLRTGY 135
GK +V G +L+VS FTL A T++G +P FS PD+A +Y+ F+E R
Sbjct 59 EGK-MNLNVQQAGGSVLVVSQFTLAADTERGMRPSFSKGASPDRAEALYDYFVERCRQ-- 115
Query 136 KPSKVQCGQFQTRMRVEIHNDGPVTL 161
+ Q G+F M+V + NDGPVT
Sbjct 116 QEMNTQTGRFAADMQVSLVNDGPVTF 141
> tpv:TP01_0031 glutamyl-tRNA amidotransferase subunit B
Length=677
Score = 31.6 bits (70), Expect = 1.6, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 17/33 (51%), Gaps = 3/33 (9%)
Query 109 PDFSAAMPPDKARPVYESF---LENLRTGYKPS 138
PD +P D R VY F LE + + YKPS
Sbjct 127 PDLYKTLPADNRRKVYNGFKEVLEEISSNYKPS 159
Lambda K H
0.317 0.134 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4535951560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40