bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2732_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
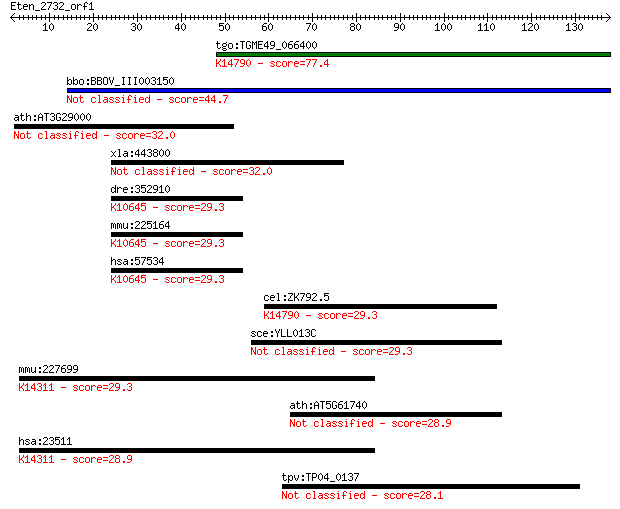
tgo:TGME49_066400 hypothetical protein ; K14790 nucleolar prot... 77.4 1e-14
bbo:BBOV_III003150 17.m07302; pumilio-family RNA binding repea... 44.7 1e-04
ath:AT3G29000 calcium-binding EF hand family protein 32.0 0.58
xla:443800 mrpl17, MGC83084; mitochondrial ribosomal protein L17 32.0 0.66
dre:352910 mib, KIAA1323, cg5841, chunp6889, fe47f05, im:71481... 29.3 3.5
mmu:225164 Mib1, DIP-1, E430019M12Rik, MGC18948, Mib, mKIAA132... 29.3 3.6
hsa:57534 MIB1, DIP-1, DKFZp686I0769, DKFZp761M1710, FLJ90676,... 29.3 3.6
cel:ZK792.5 hypothetical protein; K14790 nucleolar protein 9 29.3
sce:YLL013C PUF3; Protein of the mitochondrial outer surface, ... 29.3 4.2
mmu:227699 Nup188, BC025526, KIAA0169, U89435, mKIAA0169; nucl... 29.3 4.3
ath:AT5G61740 ATATH14; ATPase, coupled to transmembrane moveme... 28.9 4.8
hsa:23511 NUP188, FLJ21639, KIAA0169; nucleoporin 188kDa; K143... 28.9 4.8
tpv:TP04_0137 hypothetical protein 28.1 9.1
> tgo:TGME49_066400 hypothetical protein ; K14790 nucleolar protein
9
Length=960
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 41/90 (45%), Positives = 58/90 (64%), Gaps = 0/90 (0%)
Query 48 ELVKFAEALSESACGSRTLEVFFSFFSSQTFQKFFSSFLLKRAPQLAAGRFGNFLLQKVL 107
E+++ A+AL +S GSR LEV + + F+ FFSS+LLKR LA GRFGNF+LQK+L
Sbjct 411 EMMQLADALVDSPTGSRALEVAALMLAPEPFELFFSSWLLKRVNHLAQGRFGNFMLQKLL 470
Query 108 QSPSFEAPHLKLLLQALDFKEILFCGTGAV 137
S + H++ L+ A+DF + T AV
Sbjct 471 TSTLLQPAHVRQLVGAVDFSACVAARTPAV 500
> bbo:BBOV_III003150 17.m07302; pumilio-family RNA binding repeat
domain containing protein
Length=498
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/126 (25%), Positives = 58/126 (46%), Gaps = 10/126 (7%)
Query 14 CFIFFNSLFAAAAEPLQQLVEALFSSPKEKDSDEELVKFAEALSESACGSRTLEVFFSFF 73
C F A+ +EPL+ F++ E D L + S S ++
Sbjct 116 CCTLFEHQGASISEPLR-----YFATTVE---DRCLFELLHHTSGSHVLRSLMKALVGVL 167
Query 74 SSQTFQK--FFSSFLLKRAPQLAAGRFGNFLLQKVLQSPSFEAPHLKLLLQALDFKEILF 131
++ FQK + + +L A ++A FGN++LQ V ++ F+ HL+ LL+ +D +L
Sbjct 168 DAEAFQKAKLWKTSILPNAKEIAENSFGNYVLQAVTRNKYFQPTHLEELLETIDMGNLLL 227
Query 132 CGTGAV 137
+ +V
Sbjct 228 TSSSSV 233
> ath:AT3G29000 calcium-binding EF hand family protein
Length=194
Score = 32.0 bits (71), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Query 2 LFTRFNFFFVSFC-FIFFNSLFAAAAEPLQQLVEALFSSPKEKDSDEELVK 51
LF FNFF +SFC ++ +F + PL Q + +F KD E L K
Sbjct 12 LFALFNFFLISFCRWVSSTRIFLSRFVPLLQHHQRVFDKKNNKDQQETLTK 62
> xla:443800 mrpl17, MGC83084; mitochondrial ribosomal protein
L17
Length=168
Score = 32.0 bits (71), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 26/54 (48%), Gaps = 1/54 (1%)
Query 24 AAAEPLQQLVEALFSSPKEKDSDEELVKFAE-ALSESACGSRTLEVFFSFFSSQ 76
A AE LQQ E L K D+DE +K A+ L+E + +V FSS
Sbjct 47 ARAEELQQYAEKLIDYGKRGDTDERAMKMADFWLTEKDLIPKLFKVLVPRFSSH 100
> dre:352910 mib, KIAA1323, cg5841, chunp6889, fe47f05, im:7148100,
mib1, wu:fe47f05; mind bomb (EC:6.3.2.-); K10645 E3 ubiquitin-protein
ligase mind-bomb [EC:6.3.2.19]
Length=1030
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 24 AAAEPLQQLVEALFSSPKEKDSDEELVKFA 53
A+ E L QL++ LF + + D +EELVK A
Sbjct 410 ASGERLSQLLKKLFETQESGDINEELVKAA 439
> mmu:225164 Mib1, DIP-1, E430019M12Rik, MGC18948, Mib, mKIAA1323;
mindbomb homolog 1 (Drosophila); K10645 E3 ubiquitin-protein
ligase mind-bomb [EC:6.3.2.19]
Length=1006
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 24 AAAEPLQQLVEALFSSPKEKDSDEELVKFA 53
A+ E L QL++ LF + + D +EELVK A
Sbjct 410 ASGERLSQLLKKLFETQESGDLNEELVKAA 439
> hsa:57534 MIB1, DIP-1, DKFZp686I0769, DKFZp761M1710, FLJ90676,
MGC129659, MGC129660, MIB, ZZANK2, ZZZ6; mindbomb homolog
1 (Drosophila); K10645 E3 ubiquitin-protein ligase mind-bomb
[EC:6.3.2.19]
Length=1006
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 24 AAAEPLQQLVEALFSSPKEKDSDEELVKFA 53
A+ E L QL++ LF + + D +EELVK A
Sbjct 410 ASGERLSQLLKKLFETQESGDLNEELVKAA 439
> cel:ZK792.5 hypothetical protein; K14790 nucleolar protein 9
Length=630
Score = 29.3 bits (64), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 23/53 (43%), Gaps = 0/53 (0%)
Query 59 SACGSRTLEVFFSFFSSQTFQKFFSSFLLKRAPQLAAGRFGNFLLQKVLQSPS 111
S+ GSR + S + F K +L RF NF LQK++ S +
Sbjct 353 SSNGSRVWDKLMETCSEDARSLLWIEFCSKNVDELTDNRFSNFPLQKMINSST 405
> sce:YLL013C PUF3; Protein of the mitochondrial outer surface,
links the Arp2/3 complex with the mitochore during anterograde
mitochondrial movement; also binds to and promotes degradation
of mRNAs for select nuclear-encoded mitochondrial proteins
Length=879
Score = 29.3 bits (64), Expect = 4.2, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 28/57 (49%), Gaps = 1/57 (1%)
Query 56 LSESACGSRTLEVFFSFFSSQTFQKFFSSFLLKRAPQLAAGRFGNFLLQKVLQSPSF 112
LS + G R ++ F SS+ + + L P L ++GN+++Q VLQ F
Sbjct 690 LSTHSYGCRVIQRLLEFGSSEDQESILNE-LKDFIPYLIQDQYGNYVIQYVLQQDQF 745
> mmu:227699 Nup188, BC025526, KIAA0169, U89435, mKIAA0169; nucleoporin
188; K14311 nuclear pore complex protein Nup188
Length=1759
Score = 29.3 bits (64), Expect = 4.3, Method: Composition-based stats.
Identities = 24/82 (29%), Positives = 33/82 (40%), Gaps = 1/82 (1%)
Query 3 FTRFNFFF-VSFCFIFFNSLFAAAAEPLQQLVEALFSSPKEKDSDEELVKFAEALSESAC 61
+NF F +SF F+S A + L V + E D +E + A LS A
Sbjct 1593 LAEYNFLFALSFTTPTFDSEVAPSFGTLLATVNVALNMLGELDKKKESLTQAVGLSTQAE 1652
Query 62 GSRTLEVFFSFFSSQTFQKFFS 83
G+RTL+ F F S
Sbjct 1653 GTRTLKSLLMFTMENCFYLLIS 1674
> ath:AT5G61740 ATATH14; ATPase, coupled to transmembrane movement
of substances / transporter
Length=848
Score = 28.9 bits (63), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 65 TLEVFFSFFSSQTFQKFFSSFLLKRAPQLAAGRFGNFLLQKVLQSPSF 112
L++ F+F +S F K ++ ++ A+G G FL ++L+SP+F
Sbjct 353 NLQISFAFLASSIFSKVKTATVVAYTLVFASGLLGMFLFGELLESPTF 400
> hsa:23511 NUP188, FLJ21639, KIAA0169; nucleoporin 188kDa; K14311
nuclear pore complex protein Nup188
Length=1749
Score = 28.9 bits (63), Expect = 4.8, Method: Composition-based stats.
Identities = 24/82 (29%), Positives = 33/82 (40%), Gaps = 1/82 (1%)
Query 3 FTRFNFFF-VSFCFIFFNSLFAAAAEPLQQLVEALFSSPKEKDSDEELVKFAEALSESAC 61
+NF F +SF F+S A + L V + E D +E + A LS A
Sbjct 1583 LAEYNFLFALSFTTPTFDSEVAPSFGTLLATVNVALNMLGELDKKKEPLTQAVGLSTQAE 1642
Query 62 GSRTLEVFFSFFSSQTFQKFFS 83
G+RTL+ F F S
Sbjct 1643 GTRTLKSLLMFTMENCFYLLIS 1664
> tpv:TP04_0137 hypothetical protein
Length=525
Score = 28.1 bits (61), Expect = 9.1, Method: Composition-based stats.
Identities = 11/68 (16%), Positives = 32/68 (47%), Gaps = 0/68 (0%)
Query 63 SRTLEVFFSFFSSQTFQKFFSSFLLKRAPQLAAGRFGNFLLQKVLQSPSFEAPHLKLLLQ 122
S LE ++ + F+ + L + N+++Q +++P F++ HL+ ++
Sbjct 240 SYILEACLKCSTNYEYTYIFNELISNEFQTLCENNYSNYVVQAFIENPHFQSSHLQYMIH 299
Query 123 ALDFKEIL 130
++ ++
Sbjct 300 NMNIVSLM 307
Lambda K H
0.329 0.140 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40