bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2752_orf6
Length=417
Score E
Sequences producing significant alignments: (Bits) Value
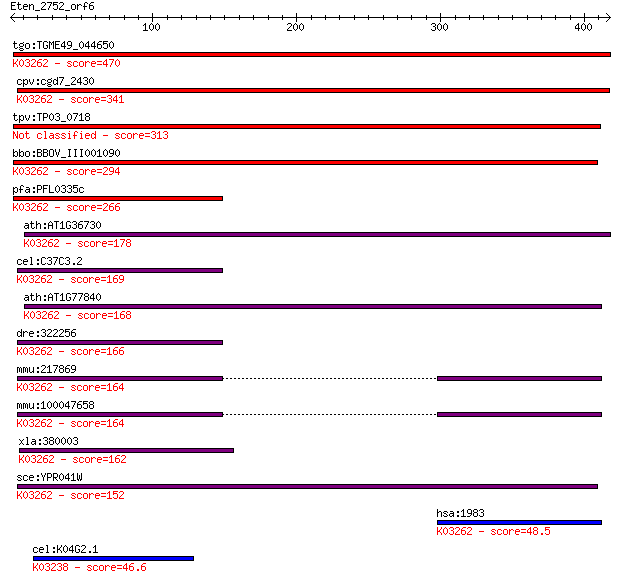
tgo:TGME49_044650 eukaryotic translation initiation factor 5, ... 470 4e-132
cpv:cgd7_2430 translation initiation factor eIF-5; Tif5p, ZnR+... 341 2e-93
tpv:TP03_0718 eukaryotic translation initiation factor 5 313 1e-84
bbo:BBOV_III001090 17.m07123; eukaryotic translation initiatio... 294 4e-79
pfa:PFL0335c eukaryotic translation initiation factor 5, putat... 266 1e-70
ath:AT1G36730 eukaryotic translation initiation factor 5, puta... 178 4e-44
cel:C37C3.2 hypothetical protein; K03262 translation initiatio... 169 3e-41
ath:AT1G77840 eukaryotic translation initiation factor 5, puta... 168 5e-41
dre:322256 eif5, fb37c12, fb54h04, wu:fb37c12, wu:fb54h04, zgc... 166 1e-40
mmu:217869 Eif5, 2810011H21Rik, D12Ertd549e, MGC36374, MGC3650... 164 8e-40
mmu:100047658 eukaryotic translation initiation factor 5-like;... 164 8e-40
xla:380003 eif5, MGC52788; eukaryotic translation initiation f... 162 3e-39
sce:YPR041W TIF5, SUI5; Tif5p; K03262 translation initiation f... 152 2e-36
hsa:1983 EIF5, EIF-5A; eukaryotic translation initiation facto... 48.5 5e-05
cel:K04G2.1 iftb-1; eIFTwoBeta (eIF2beta translation initiatio... 46.6 2e-04
> tgo:TGME49_044650 eukaryotic translation initiation factor 5,
putative ; K03262 translation initiation factor 5
Length=414
Score = 470 bits (1210), Expect = 4e-132, Method: Compositional matrix adjust.
Identities = 261/418 (62%), Positives = 319/418 (76%), Gaps = 9/418 (2%)
Query 3 MQYVNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGC 62
M VNIPRDR+DPNYRYKMPKL+SKIEGRGNGI+TN++NMGEIARALKRPPMYPTKFFGC
Sbjct 1 MALVNIPRDRDDPNYRYKMPKLVSKIEGRGNGIKTNIFNMGEIARALKRPPMYPTKFFGC 60
Query 63 ELGAMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKGVLVCK 122
ELGAM KFEE EEKAL+NGAH E DLV ILDKFIQMYVLC GC+LPEID+ VKKG+L CK
Sbjct 61 ELGAMAKFEEAEEKALVNGAHKESDLVNILDKFIQMYVLCPGCELPEIDLIVKKGLLTCK 120
Query 123 CNACGYSGTLDNTHKAATYMAKNPPNATETTLGKKKKTKEERRAEKHAKEEDKSEKTGKE 182
CNACGY G+LDN HKAATYM +NPP+A +T+GKKKK+KEERRAEK ++K + GKE
Sbjct 121 CNACGYQGSLDNVHKAATYMVRNPPDAGSSTMGKKKKSKEERRAEK----QEKQAREGKE 176
Query 183 KKSKDKDKKKKKGEEENDTDCLNSKAVDHSGDATPTNAGE-DSWDEEKSSEADKKEKKKK 241
KK K+K KKK ++++D D N K + SGDATPT+ E DSWD+EK+ + +KEKKKK
Sbjct 177 KKEKEKKKKKADSDDDSDDDGQNGK--EASGDATPTHDAETDSWDDEKTEKEKRKEKKKK 234
Query 242 EKKHANA--EGPKLVVKEALTINCPEIAEVAARLRNMIVTSSAVTPDSFFVELRMLQVSQ 299
+KK ++A KL+ KEAL + PE+ +V RLRN++ S V +FF E+RMLQVSQ
Sbjct 235 DKKKSSALDSEKKLMHKEALVFDSPELKDVIERLRNVVTRSETVDLAAFFHEMRMLQVSQ 294
Query 300 DFDAKCRMYVVLSALFKDGLSPELLEEKMAFVAKCCDSSVHASDLLSALENFCFENPHTA 359
DFD+KCR+YV L+A+F D ++P LE +M ++ K D+SV ASD+L A +C E TA
Sbjct 295 DFDSKCRIYVTLAAIFNDDMTPTSLEARMPYILKVLDTSVSASDVLDAFGFYCQEKGGTA 354
Query 360 MAGYPYMLQRLYNAELLEPEDILAYYNSPRADAATQKCRTFAEPFLKWLAEADSSDEE 417
M +PY LQ+LYNAE LE EDIL YY + + D C+ AEPFL+WLAE D S EE
Sbjct 355 MTSFPYCLQKLYNAEALEAEDILKYYAADKEDPVFSACKKQAEPFLQWLAEDDGSSEE 412
> cpv:cgd7_2430 translation initiation factor eIF-5; Tif5p, ZnR+W2
domains ; K03262 translation initiation factor 5
Length=460
Score = 341 bits (875), Expect = 2e-93, Method: Compositional matrix adjust.
Identities = 186/427 (43%), Positives = 255/427 (59%), Gaps = 59/427 (13%)
Query 6 VNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 65
+NIP +DPNYRYKMPKL+SKIEGRGNGIRT + NM +IAR+LKRPP YPTKFFGCELG
Sbjct 2 LNIPSTVDDPNYRYKMPKLVSKIEGRGNGIRTKIVNMADIARSLKRPPAYPTKFFGCELG 61
Query 66 AMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKGVLVCKCNA 125
A+ K+E+ EEK+++NGAH + DL ++DKFIQMYVLC C+LPEIDI VKK + CKCNA
Sbjct 62 ALSKWEDKEEKSIVNGAHQQNDLQRLMDKFIQMYVLCPNCELPEIDIIVKKERVSCKCNA 121
Query 126 CGYSGTLDNTHKAATYMAKNPPNATETTLGKKKKTKEERRAEKHAKEEDKSEKTGKEKKS 185
CGY G LDN HK TY++KNPP E+T+G K K KE++
Sbjct 122 CGYIGALDNNHKITTYISKNPPGGVESTMGSKNK---------------------KERRE 160
Query 186 KDKDKKKKKGEEENDTDCLNSKAVDHSGDATPTNAGEDSWDEEKSSEADKKEKKKKEKKH 245
+DKD+ + C NS G + D EK S+ K+ KK K
Sbjct 161 RDKDRSSEG--------CNNSN-------------GSTAADNEKPSKHKSKKASKKSSKK 199
Query 246 ANAEGPKLVVKEALTINCPEIAEVAARLRNMIVTSSAVTPDSFFVELRMLQVSQDFDAKC 305
+ E + K+ LT PEI +V RL N + + V+P+ FF ELR+LQVSQDFD
Sbjct 200 SPHE-LDIFEKDVLTFESPEIVDVIERLTNFMQKNEEVSPEDFFSELRVLQVSQDFDEFM 258
Query 306 RMYVVLSALF--KDGLSPELLEEKMAFVAKCCDSSVHASDLLSALENFCFENPHTAMAGY 363
R V+L+ LF KD + P++ + ++ +++K + S+ ++ ++S E++C E + +A Y
Sbjct 259 RFSVLLAVLFGLKDPIDPKVFKLRIPYISKAVEGSLRSNAIISIFESYCVEKNPSTLATY 318
Query 364 PYMLQRLYNAELLEPEDILAYYNSPRADAATQKCRT---------FAEPFLKWLA----- 409
PY++Q+LY+A++L + IL YY + C T EPF+ WL
Sbjct 319 PYLIQQLYDADILSEKVILKYYQKDAPTSWVSGCSTQDTFEKAKKSCEPFVDWLLDGSND 378
Query 410 EADSSDE 416
E+D SDE
Sbjct 379 ESDLSDE 385
> tpv:TP03_0718 eukaryotic translation initiation factor 5
Length=385
Score = 313 bits (801), Expect = 1e-84, Method: Compositional matrix adjust.
Identities = 179/415 (43%), Positives = 240/415 (57%), Gaps = 39/415 (9%)
Query 3 MQYVNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGC 62
M YVNIPR R+DPNYRYKMP++ S+IEGRGNG +TN+ NMG+IARALKRPP Y TKFFGC
Sbjct 1 MSYVNIPRYRDDPNYRYKMPRIQSRIEGRGNGTKTNISNMGDIARALKRPPTYATKFFGC 60
Query 63 ELGAMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKGVLVCK 122
ELGAM KFEE+EEKALINGAH++ L +LDKFI++YVLC C LPEI++ VK+G L+C
Sbjct 61 ELGAMSKFEESEEKALINGAHTDTTLAGVLDKFIELYVLCQNCQLPEIELFVKRGELLCS 120
Query 123 CNACGYSGTLDNTHKAATYMAKNPPNATETTLGKKKKTKEERRAEKHAKEEDKSEKTGKE 182
CNACG+ GTLD THKAA+YM KNP N K TKE +++ + E
Sbjct 121 CNACGHKGTLDMTHKAASYMIKNPINTA-------KMTKEGSSSKEPTNGVANNTVVKSE 173
Query 183 KKSKDKDKKKKKGEEENDTDCLNSKAVDHSGDATPTNAGEDSWDEEKSSEADKKEKKKKE 242
K K +D K + E++D + + E S EKS + DK EK
Sbjct 174 KSKKHRDPSKSEKNEKSDKS-------EKPEKVEKSEKTEKSEKSEKSEKIDKVEKL--- 223
Query 243 KKHANAEGPKLVVKEALTINCPEIAEVAARLRNMIVTSSAVTPDSFFVELRMLQVSQDFD 302
E L++ PE++E+A RLR + D F EL MLQ+SQ FD
Sbjct 224 --------------EDLSVESPELSEIATRLRCFLEKGDRTCED-FSEELHMLQISQGFD 268
Query 303 AKCRMYVVLSALFKDG--LSPELLEEKMAFVAKCCDSSVHASDLLSALENFCFENPHTAM 360
RM++ LSA+F L+ EL ++++ V S+ + ++ALE F F++ +
Sbjct 269 NVSRMFIALSAIFNSDVPLTLELFKDRIQLVKAATGYSMTPKEKITALEYFIFQSNPQEI 328
Query 361 AGYPYMLQRLYNAELLEPEDILAYYN-----SPRADAATQKCRTFAEPFLKWLAE 410
YPY Q LY+ ++LE D + YYN S + + + PF++WL E
Sbjct 329 TNYPYYSQLLYSHDILEGSDFIRYYNKSNRHSGNLNNIFLRAKETIAPFIEWLQE 383
> bbo:BBOV_III001090 17.m07123; eukaryotic translation initiation
factor 5; K03262 translation initiation factor 5
Length=379
Score = 294 bits (753), Expect = 4e-79, Method: Compositional matrix adjust.
Identities = 171/411 (41%), Positives = 221/411 (53%), Gaps = 50/411 (12%)
Query 3 MQYVNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGC 62
M VNIPR EDPNYRYKMPKL SKIEGRGNGI+TN+ NMG+IARALKRPP Y TKFFGC
Sbjct 1 MSLVNIPRYCEDPNYRYKMPKLQSKIEGRGNGIKTNISNMGDIARALKRPPTYATKFFGC 60
Query 63 ELGAMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKGVLVCK 122
ELGAM KFEE+EEKALINGAH+E L ILDKFI+MYVLC C LPEI++ VK+G L +
Sbjct 61 ELGAMSKFEESEEKALINGAHTERALTQILDKFIEMYVLCAECHLPEIEVLVKRGALCSR 120
Query 123 CNACGYSGTLDNTHKAATYMAKNPPNATETTLGKKKKTKEERRAEKHAKEEDKSEKTGKE 182
CNACGY G LDNTHKAA YM KNP TE
Sbjct 121 CNACGYKGVLDNTHKAAGYMLKNPACLTEM------------------------------ 150
Query 183 KKSKDKDKKKKKGEEENDTDCLNSKAVDHSGDATPTNAGEDSWDEEKSSEADKKEKKKKE 242
SK ++ KK+ ++ +T+ K + S D ++ + E
Sbjct 151 --SKHSNRTKKESTKDGNTEKDKDKEKEKKKKDKKEKKKRISDDGQELINGGDGATQTME 208
Query 243 KKHANAEGPKLVVKEALTINCPEIAEVAARLRNMIVTSSAVTPDSFFVELRMLQVSQDFD 302
+ E P EI + R+ + + P F EL MLQ+SQDFD
Sbjct 209 MEKLTIESP-------------EIQDAIERITSSLKGGKFQQPAEFAEELHMLQLSQDFD 255
Query 303 AKCRMYVVLSALFKDGLSPELLEEKMAFVAKCCDSSVHASDLLSALENFCFENPHTAMAG 362
C++YV L A+F D + LE+K+ + + V D+L+++ F + +
Sbjct 256 NICKVYVALCAIFSDKFDVQELEDKINLLIPLLEYGVQPKDVLTSMAVFVVKLYPQQLDD 315
Query 363 YPYMLQRLYNAELLEPEDILAYY-NSPRADA----ATQKCRTFAEPFLKWL 408
YPY+ + LY ++L + DI +Y S R D A + + AEPF+ WL
Sbjct 316 YPYICRALYASDLADGGDICRFYAKSTRFDGSLAEAYIRTKEKAEPFVAWL 366
> pfa:PFL0335c eukaryotic translation initiation factor 5, putative;
K03262 translation initiation factor 5
Length=565
Score = 266 bits (679), Expect = 1e-70, Method: Compositional matrix adjust.
Identities = 118/145 (81%), Positives = 129/145 (88%), Gaps = 0/145 (0%)
Query 3 MQYVNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGC 62
M YVNIPRDR DPNYRYKMPKL+SKIEGRGNGIRTN+ NMGEIAR+LKRPPMYPTKFFGC
Sbjct 1 MSYVNIPRDRNDPNYRYKMPKLISKIEGRGNGIRTNISNMGEIARSLKRPPMYPTKFFGC 60
Query 63 ELGAMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKGVLVCK 122
ELG MVKFEENEEKA++NGAH E+DLV ILDKFI+MYVLC C LPE DI VKKG+L+CK
Sbjct 61 ELGTMVKFEENEEKAIVNGAHKEKDLVNILDKFIEMYVLCPHCLLPETDIVVKKGILICK 120
Query 123 CNACGYSGTLDNTHKAATYMAKNPP 147
CNACG G L+N+HK ATYM KNPP
Sbjct 121 CNACGNIGELNNSHKIATYMIKNPP 145
> ath:AT1G36730 eukaryotic translation initiation factor 5, putative
/ eIF-5, putative; K03262 translation initiation factor
5
Length=439
Score = 178 bits (451), Expect = 4e-44, Method: Compositional matrix adjust.
Identities = 142/438 (32%), Positives = 219/438 (50%), Gaps = 40/438 (9%)
Query 11 DREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKF 70
+R+D YRYKMP++++KIEGRGNGI+TNV NM EIA+AL RP Y TK+FGCELGA KF
Sbjct 10 NRDDAFYRYKMPRMMTKIEGRGNGIKTNVVNMVEIAKALGRPAAYTTKYFGCELGAQSKF 69
Query 71 EENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKG-VLVCKCNACGYS 129
+E +L+NGAH L +L+ FI+ YV C GC PE +I + K +L KC ACG+
Sbjct 70 DEKNGTSLVNGAHDTSKLAGLLENFIKKYVQCYGCGNPETEILITKTQMLQLKCAACGFL 129
Query 130 GTLDNTHKAATYMAKNPPNATETTLGK---KKKTKEERRAEKHAKEEDKSEKTGKEKKSK 186
+D K +++ KNPP +++ K ++ KE R + A EE + K K K
Sbjct 130 SDVDMRDKLTSFILKNPPEQKKSSKDKKSMRRAEKERLREGEAADEEMRKLKKEAASKKK 189
Query 187 DKDKKKKKGEEENDTDCLNSKAVDHSGDATPTNAGED--SWDEEKSSEADKKEKKKK--- 241
K + D +++ D + +D W + S EA +K K++
Sbjct 190 AATTGTSKDKVSKKKDHSPPRSLSDENDQADSEEDDDDVQWQTDTSREAAEKRMKEQLSA 249
Query 242 -----------EKKHANAEGPKL--VVKEALTINCPEIAEVAARLRNMI--VTSSAVTPD 286
E+K AE K V E PE A +L N I + SS +P
Sbjct 250 VTAEMVMLSTVEEKKPVAEVKKAPEQVHENGNSKIPENAH--EKLVNEIKELLSSGSSP- 306
Query 287 SFFVELRMLQVSQDFDAKCRMYVVLSALFKD---GLSPELLEEK---MAFVAKCCDSSVH 340
+L+ S + + +M + SALF G + E++++K +A + ++
Sbjct 307 ---TQLKTALASNSANPQEKMDALFSALFGGTGKGFAKEVIKKKKYLLALMMMQEEAGAP 363
Query 341 AS-DLLSALENFCFENPHTAMAGYPYMLQRLYNAELLEPEDILAYYNSPRADAATQKCRT 399
A LL+ +E+FC + A +++ LY+ ++L+ + I+ +YN + K T
Sbjct 364 AQMGLLNGIESFCMKASAEAAKEVALVIKGLYDEDILDEDVIVEWYNKGVKSSPVLKNVT 423
Query 400 FAEPFLKWLAEADSSDEE 417
PF++WL A+S EE
Sbjct 424 ---PFIEWLQNAESESEE 438
> cel:C37C3.2 hypothetical protein; K03262 translation initiation
factor 5
Length=413
Score = 169 bits (427), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 75/142 (52%), Positives = 99/142 (69%), Gaps = 0/142 (0%)
Query 6 VNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 65
+N+ R DP YRYKMPKL +K+EG+GNGI+T + NM EIA+AL+RPPMYPTK+FGCELG
Sbjct 3 LNVNRAVADPFYRYKMPKLSAKVEGKGNGIKTVISNMSEIAKALERPPMYPTKYFGCELG 62
Query 66 AMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKGVLVCKCNA 125
A F+ E+ ++NG H L ILD FI+ +VLC C+ PE + V+K + KC A
Sbjct 63 AQTNFDAKNERYIVNGEHDANKLQDILDGFIKKFVLCKSCENPETQLFVRKNNIKSKCKA 122
Query 126 CGYSGTLDNTHKAATYMAKNPP 147
CG S +D HK +T++ KNPP
Sbjct 123 CGCSFDIDLKHKLSTFIMKNPP 144
> ath:AT1G77840 eukaryotic translation initiation factor 5, putative
/ eIF-5, putative; K03262 translation initiation factor
5
Length=437
Score = 168 bits (425), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 129/434 (29%), Positives = 211/434 (48%), Gaps = 47/434 (10%)
Query 11 DREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKF 70
+R D YRYKMPK+++K EG+GNGI+TN+ N EIA+AL RPP Y TK+FGCELGA KF
Sbjct 10 NRNDAFYRYKMPKMVTKTEGKGNGIKTNIINNVEIAKALARPPSYTTKYFGCELGAQSKF 69
Query 71 EENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKGVLV-CKCNACGYS 129
+E +L+NGAH+ L +L+ FI+ +V C GC PE +I + K +V KC ACG+
Sbjct 70 DEKTGTSLVNGAHNTSKLAGLLENFIKKFVQCYGCGNPETEIIITKTQMVNLKCAACGFI 129
Query 130 GTLDNTHKAATYMAKNPPNATETTLGKK--KKTKEERRAEKH-AKEEDKSEKTGKEKKSK 186
+D K ++ KNPP + + KK +K ++ER E A EE + K K+ S
Sbjct 130 SEVDMRDKLTNFILKNPPEQKKVSKDKKAMRKAEKERLKEGELADEEQRKLKAKKKALSN 189
Query 187 DKDKKKKKGEEENDTDCLNSKAVDHSGDATPTNAGEDSWDEEKSSEADKKEKKKKEKKHA 246
KD K K ++ D + W + S EA +K+ +
Sbjct 190 GKDSKTSKNHSSDEDISPKHDENALEVDEDEDDDDGVEWQTDTSREA----AEKRMMEQL 245
Query 247 NAEGPKLVVKEALTIN-------------------CPEIAEVAARLRNMIVTSSAVTPDS 287
+A+ ++V+ A+ + P+ + ++ + S ++
Sbjct 246 SAKTAEMVMLSAMEVEEKKAPKSKSNGNVVKTENPPPQEKNLVQDMKEYLKKGSPISALK 305
Query 288 FFVELRMLQVSQDFDAKCRMYVVLSALFKD---GLSPELLEEKMAFVAKCC----DSSVH 340
F+ + + QD M + +ALF G + E+ ++K A S +H
Sbjct 306 SFIS-SLSEPPQDI-----MDALFNALFDGVGKGFAKEVTKKKNYLAAAATMQEDGSQMH 359
Query 341 ASDLLSALENFCFENPH-TAMAGYPYMLQRLYNAELLEPEDILAYYNS--PRADAATQKC 397
LL+++ FC +N + A+ +L+ LY+ +++E E +L +Y AD ++
Sbjct 360 ---LLNSIGTFCGKNGNEEALKEVALVLKALYDQDIIEEEVVLDWYEKGLTGADKSSPVW 416
Query 398 RTFAEPFLKWLAEA 411
+ +PF++WL A
Sbjct 417 KN-VKPFVEWLQSA 429
> dre:322256 eif5, fb37c12, fb54h04, wu:fb37c12, wu:fb54h04, zgc:56606,
zgc:77026; eukaryotic translation initiation factor
5; K03262 translation initiation factor 5
Length=429
Score = 166 bits (421), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 72/144 (50%), Positives = 99/144 (68%), Gaps = 2/144 (1%)
Query 6 VNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 65
+N+ R D YRYKMP+L++K+EG+GNGI+T + NM ++A+AL RPP YPTKFFGCELG
Sbjct 3 INVNRSVSDQFYRYKMPRLIAKVEGKGNGIKTVIVNMVDVAKALNRPPTYPTKFFGCELG 62
Query 66 AMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITV--KKGVLVCKC 123
A +F+ ++ ++NG+H L +LD FI+ +VLC CD PE D+ + KK + C
Sbjct 63 AQTQFDSKNDRYIVNGSHEANKLQDMLDGFIRKFVLCPECDNPETDLHINPKKQTIGNSC 122
Query 124 NACGYSGTLDNTHKAATYMAKNPP 147
ACGY G LD HK T++ KNPP
Sbjct 123 KACGYRGMLDTRHKLCTFILKNPP 146
> mmu:217869 Eif5, 2810011H21Rik, D12Ertd549e, MGC36374, MGC36509;
eukaryotic translation initiation factor 5; K03262 translation
initiation factor 5
Length=429
Score = 164 bits (414), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 72/144 (50%), Positives = 99/144 (68%), Gaps = 2/144 (1%)
Query 6 VNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 65
VN+ R D YRYKMP+L++K+EG+GNGI+T + NM ++A+AL RPP YPTK+FGCELG
Sbjct 3 VNVNRSVSDQFYRYKMPRLIAKVEGKGNGIKTVIVNMVDVAKALNRPPTYPTKYFGCELG 62
Query 66 AMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITV--KKGVLVCKC 123
A +F+ ++ ++NG+H L +LD FI+ +VLC C+ PE D+ V KK + C
Sbjct 63 AQTQFDVKNDRYIVNGSHEANKLQDMLDGFIKKFVLCPECENPETDLHVNPKKQTIGNSC 122
Query 124 NACGYSGTLDNTHKAATYMAKNPP 147
ACGY G LD HK T++ KNPP
Sbjct 123 KACGYRGMLDTHHKLCTFILKNPP 146
Score = 48.1 bits (113), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 34/121 (28%), Positives = 62/121 (51%), Gaps = 9/121 (7%)
Query 298 SQDFDAKCRMYVVLS-ALFKDGLSPELLEEKMAFVAKCCDSSVHASDLLSALENFCFENP 356
++ D K +VL+ LF + + ++ + + F+ C ++ LL LE C
Sbjct 265 AERLDVKAMGPLVLTEVLFDEKIREQIKKYRRHFLRFCHNNKKAQRYLLHGLE--CVVAM 322
Query 357 HTA--MAGYPYMLQRLYNAELLEPEDILAYYNSPRADAAT----QKCRTFAEPFLKWLAE 410
H A ++ P++L+ +Y+A+LLE E I+++ + ++ R AEPF+KWL E
Sbjct 323 HQAQLISKIPHILKEMYDADLLEEEVIISWSEKASKKYVSKELAKEIRVKAEPFIKWLKE 382
Query 411 A 411
A
Sbjct 383 A 383
> mmu:100047658 eukaryotic translation initiation factor 5-like;
K03262 translation initiation factor 5
Length=429
Score = 164 bits (414), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 72/144 (50%), Positives = 99/144 (68%), Gaps = 2/144 (1%)
Query 6 VNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 65
VN+ R D YRYKMP+L++K+EG+GNGI+T + NM ++A+AL RPP YPTK+FGCELG
Sbjct 3 VNVNRSVSDQFYRYKMPRLIAKVEGKGNGIKTVIVNMVDVAKALNRPPTYPTKYFGCELG 62
Query 66 AMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITV--KKGVLVCKC 123
A +F+ ++ ++NG+H L +LD FI+ +VLC C+ PE D+ V KK + C
Sbjct 63 AQTQFDVKNDRYIVNGSHEANKLQDMLDGFIKKFVLCPECENPETDLHVNPKKQTIGNSC 122
Query 124 NACGYSGTLDNTHKAATYMAKNPP 147
ACGY G LD HK T++ KNPP
Sbjct 123 KACGYRGMLDTHHKLCTFILKNPP 146
Score = 48.1 bits (113), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 34/121 (28%), Positives = 62/121 (51%), Gaps = 9/121 (7%)
Query 298 SQDFDAKCRMYVVLS-ALFKDGLSPELLEEKMAFVAKCCDSSVHASDLLSALENFCFENP 356
++ D K +VL+ LF + + ++ + + F+ C ++ LL LE C
Sbjct 265 AERLDVKAMGPLVLTEVLFDEKIREQIKKYRRHFLRFCHNNKKAQRYLLHGLE--CVVAM 322
Query 357 HTA--MAGYPYMLQRLYNAELLEPEDILAYYNSPRADAAT----QKCRTFAEPFLKWLAE 410
H A ++ P++L+ +Y+A+LLE E I+++ + ++ R AEPF+KWL E
Sbjct 323 HQAQLISKIPHILKEMYDADLLEEEVIISWSEKASKKYVSKELAKEIRVKAEPFIKWLKE 382
Query 411 A 411
A
Sbjct 383 A 383
> xla:380003 eif5, MGC52788; eukaryotic translation initiation
factor 5; K03262 translation initiation factor 5
Length=434
Score = 162 bits (409), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 73/151 (48%), Positives = 100/151 (66%), Gaps = 2/151 (1%)
Query 7 NIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGA 66
N+ R D YRYKMP+L++K+EG+GNGI+T + NM ++A+AL RPP YPTK+FGCELGA
Sbjct 4 NVNRSVSDQFYRYKMPRLIAKVEGKGNGIKTVIVNMVDVAKALNRPPTYPTKYFGCELGA 63
Query 67 MVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITV--KKGVLVCKCN 124
+F+ ++ ++NG+H L +LD FI+ +VLC CD PE D+ V KK + C
Sbjct 64 QTQFDIKNDRFIVNGSHEANKLQDMLDGFIKKFVLCPECDNPETDLHVNPKKQTIGNSCK 123
Query 125 ACGYSGTLDNTHKAATYMAKNPPNATETTLG 155
ACGY G LD HK T++ KNP T+ G
Sbjct 124 ACGYRGMLDTKHKLCTFILKNPTENTDGNAG 154
> sce:YPR041W TIF5, SUI5; Tif5p; K03262 translation initiation
factor 5
Length=405
Score = 152 bits (384), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 114/410 (27%), Positives = 177/410 (43%), Gaps = 27/410 (6%)
Query 6 VNIPRDREDPNYRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELG 65
+NI RD DP YRYKMP + +K+EGRGNGI+T V N+ +I+ AL RP Y K+FG ELG
Sbjct 3 INICRDNHDPFYRYKMPPIQAKVEGRGNGIKTAVLNVADISHALNRPAPYIVKYFGFELG 62
Query 66 AMVKFEENEEKALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKK-GVLVCKCN 124
A ++++ L+NG H L +LD FI +VLCG C PE +I + K LV C
Sbjct 63 AQTSISVDKDRYLVNGVHEPAKLQDVLDGFINKFVLCGSCKNPETEIIITKDNDLVRDCK 122
Query 125 ACGYSGTLDNTHKAATYMAKNPPNATETTLGKKKKTKEERRAEKHAKEEDKSEKTGKEKK 184
ACG +D HK ++++ KNPP++ + KKK + +
Sbjct 123 ACGKRTPMDLRHKLSSFILKNPPDSVSGSKKKKKAATASANVRGGGLSISDIAQGKSQNA 182
Query 185 SKDKDKKKKKGEEENDTDCLNSKAVDHSGDATPTNAGEDSWDEEKSSEADKKEKKKKEKK 244
D + D D L+ + + +D W + S EA + K+ E
Sbjct 183 PSDGTGSSTPQHHDEDEDELSRQIKAAASTLEDIEVKDDEWAVDMSEEAIRARAKELE-- 240
Query 245 HANAEGPKLVVKEALTINCPEIAEVAARLRNMIVTSSAVTPDS--FFVELRMLQVSQDFD 302
V LT ++ E + P + + L V D
Sbjct 241 ----------VNSELT----QLDEYGEWILEQAGEDKENLPSDVELYKKAAELDVLNDPK 286
Query 303 AKCRMYVVLSALFKDGLSPELLEEKMAFVAKCCDSSVHASDLLSALENFCFENPHTAMAG 362
C V+ LF + + E+ E AF K + + + + +E F +
Sbjct 287 IGC---VLAQCLFDEDIVNEIAEHN-AFFTKILVTPEYEKNFMGGIERFLGLEHKDLIPL 342
Query 363 YPYMLQRLYNAELLEPEDILAYYNSPR----ADAATQKCRTFAEPFLKWL 408
P +L +LYN +++ E+I+ + ++K R A+PF+ WL
Sbjct 343 LPKILVQLYNNDIISEEEIMRFGTKSSKKFVPKEVSKKVRRAAKPFITWL 392
> hsa:1983 EIF5, EIF-5A; eukaryotic translation initiation factor
5; K03262 translation initiation factor 5
Length=431
Score = 48.5 bits (114), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 34/121 (28%), Positives = 62/121 (51%), Gaps = 9/121 (7%)
Query 298 SQDFDAKCRMYVVLS-ALFKDGLSPELLEEKMAFVAKCCDSSVHASDLLSALENFCFENP 356
++ D K +VL+ LF + + ++ + + F+ C ++ LL LE C
Sbjct 267 AERLDVKAMGPLVLTEVLFNEKIREQIKKYRRHFLRFCHNNKKAQRYLLHGLE--CVVAM 324
Query 357 HTA--MAGYPYMLQRLYNAELLEPEDILAYYNSPRADAAT----QKCRTFAEPFLKWLAE 410
H A ++ P++L+ +Y+A+LLE E I+++ + ++ R AEPF+KWL E
Sbjct 325 HQAQLISKIPHILKEMYDADLLEEEVIISWSEKASKKYVSKELAKEIRVKAEPFIKWLKE 384
Query 411 A 411
A
Sbjct 385 A 385
> cel:K04G2.1 iftb-1; eIFTwoBeta (eIF2beta translation initiation
factor) family member (iftb-1); K03238 translation initiation
factor 2 subunit 2
Length=250
Score = 46.6 bits (109), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 49/111 (44%), Gaps = 6/111 (5%)
Query 17 YRYKMPKLLSKIEGRGNGIRTNVYNMGEIARALKRPPMYPTKFFGCELGAMVKFEENEEK 76
+ K+P++ R +T N EI R +KR + +F ELG +
Sbjct 114 FAIKLPEV-----ARAGSKKTAFSNFLEICRLMKRQDKHVLQFLLAELGTTGSID-GSNC 167
Query 77 ALINGAHSEEDLVAILDKFIQMYVLCGGCDLPEIDITVKKGVLVCKCNACG 127
++ G ++ ++L K+I+ YV+C C PE +T + +C CG
Sbjct 168 LIVKGRWQQKQFESVLRKYIKEYVMCHTCKSPETQLTKDTRLFFLQCTNCG 218
Lambda K H
0.313 0.129 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 19072613096
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40