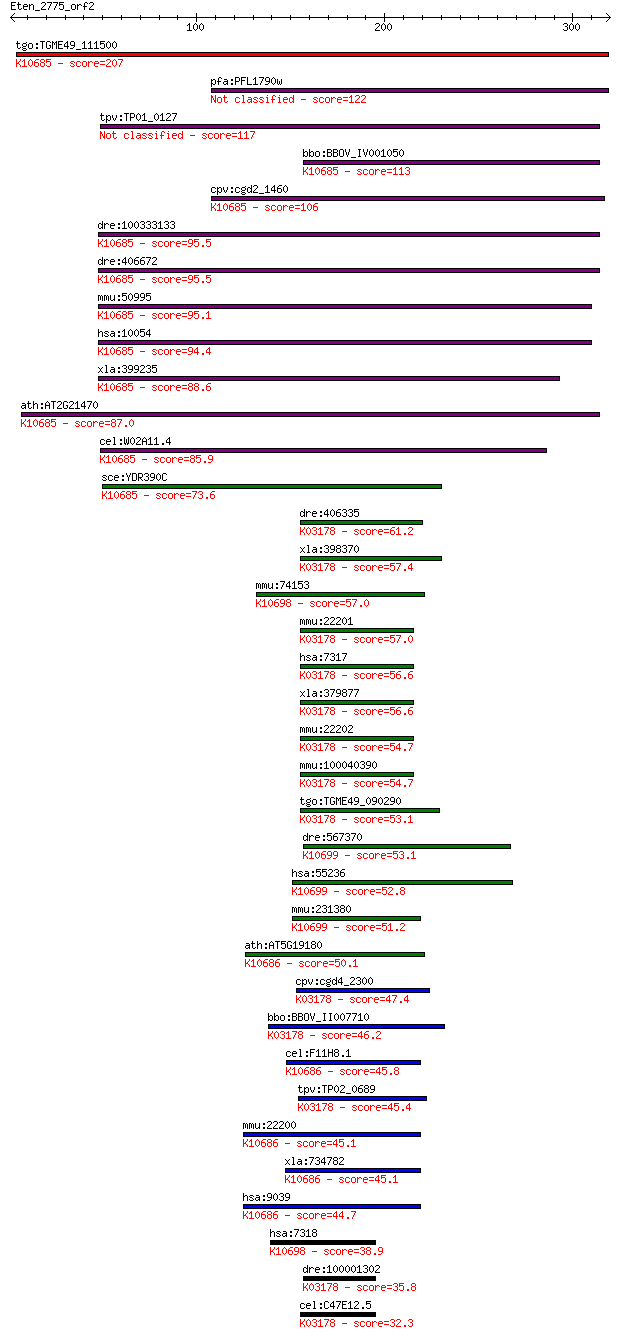
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2775_orf2
Length=319
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_111500 ubiquitin-activating enzyme, putative (EC:1.... 207 6e-53
pfa:PFL1790w ubiquitin-activating enzyme, putative 122 2e-27
tpv:TP01_0127 ubiquitin-protein ligase 117 6e-26
bbo:BBOV_IV001050 21.m02728; ubiquitin-activating enzyme; K106... 113 1e-24
cpv:cgd2_1460 SUMO-1 activating enzyme subunit 2 ; K10685 ubiq... 106 9e-23
dre:100333133 ubiquitin-like modifier activating enzyme 2-like... 95.5 2e-19
dre:406672 uba2, sae2, sae2b, uble1b, wu:fi17g06, zgc:66354; u... 95.5 2e-19
mmu:50995 Uba2, AA986091, Arx, Sae2, UBA1, Ubl1a2, Uble1b; ubi... 95.1 3e-19
hsa:10054 UBA2, ARX, FLJ13058, SAE2; ubiquitin-like modifier a... 94.4 6e-19
xla:399235 uba2, MGC84651, Uble1b, sae2, sae2-B, uba2-a, uba2-... 88.6 3e-17
ath:AT2G21470 SAE2; SAE2 (SUMO-ACTIVATING ENZYME 2); SUMO acti... 87.0 8e-17
cel:W02A11.4 uba-2; UBA (human ubiquitin) related family membe... 85.9 2e-16
sce:YDR390C UBA2, UAL1; Nuclear protein that acts as a heterod... 73.6 8e-13
dre:406335 uba1, ube1, wu:fa01e08, wu:fb30f01, wu:fi21c11, wu:... 61.2 5e-09
xla:398370 uba1-a, MGC68851, a1s9, a1s9t, a1st, amcx1, gxp1, p... 57.4 8e-08
mmu:74153 Uba7, 1300004C08Rik, Ube1l; ubiquitin-like modifier ... 57.0 8e-08
mmu:22201 Uba1, A1S9, AA989744, Sbx, Ube-1, Ube1x; ubiquitin-l... 57.0 9e-08
hsa:7317 UBA1, A1S9, A1S9T, A1ST, AMCX1, GXP1, MGC4781, POC20,... 56.6 1e-07
xla:379877 uba1-b, MGC52522, a1s9, a1s9t, a1st, amcx1, gxp1, p... 56.6 1e-07
mmu:22202 Ube1y1, A1s9Y-1, Sby, Ube-2, Ube1ay, Ube1y, Ube1y-1;... 54.7 4e-07
mmu:100040390 ubiquitin-like modifier-activating enzyme 1 Y-li... 54.7 4e-07
tgo:TGME49_090290 ubiquitin-activating enzyme E1, putative ; K... 53.1 1e-06
dre:567370 ubiquitin-activating enzyme E1-like; K10699 ubiquit... 53.1 1e-06
hsa:55236 UBA6, E1-L2, FLJ10808, FLJ23367, MOP-4, UBE1L2; ubiq... 52.8 2e-06
mmu:231380 Uba6, 4930542H01, 5730469D23Rik, AU021846, AW124799... 51.2 5e-06
ath:AT5G19180 ECR1; ECR1 (E1 C-terminal related 1); NEDD8 acti... 50.1 1e-05
cpv:cgd4_2300 ubiquitin-activating enzyme E1 (UBA) ; K03178 ub... 47.4 8e-05
bbo:BBOV_II007710 18.m06639; ubiquitin-activating enzyme E1; K... 46.2 2e-04
cel:F11H8.1 rfl-1; ectopic membrane RuFfLes in embryo family m... 45.8 2e-04
tpv:TP02_0689 ubiquitin-protein ligase; K03178 ubiquitin-activ... 45.4 3e-04
mmu:22200 Uba3, A830034N06Rik, AI256736, AI848246, AW546539, U... 45.1 4e-04
xla:734782 uba3, MGC131020, ube1c; ubiquitin-like modifier act... 45.1 4e-04
hsa:9039 UBA3, DKFZp566J164, MGC22384, UBE1C, hUBA3; ubiquitin... 44.7 5e-04
hsa:7318 UBA7, D8, MGC12713, UBA1B, UBE1L, UBE2; ubiquitin-lik... 38.9 0.027
dre:100001302 ubiquitin-like modifier-activating enzyme 1-like... 35.8 0.22
cel:C47E12.5 uba-1; UBA (human ubiquitin) related family membe... 32.3 2.7
> tgo:TGME49_111500 ubiquitin-activating enzyme, putative (EC:1.2.1.70);
K10685 ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=730
Score = 207 bits (526), Expect = 6e-53, Method: Compositional matrix adjust.
Identities = 124/353 (35%), Positives = 183/353 (51%), Gaps = 81/353 (22%)
Query 4 EDPANLLADLKQDLDALCCCSSSSSSSG-----SGSGSGSGGGGLAAAGVR--------- 49
ED NLL+DLK+ L SS++ + G S G+ G G +AG R
Sbjct 220 EDNENLLSDLKEQLRTFLDVSSTAEAEGREDETSRPGTHEKGAGEDSAGTREEAMRLMSR 279
Query 50 -VMEEVFSKQIKDLLKLKKNWKSGKE-----PQPPLIQPQLLLQWYQQQQQQQQGQQQQQ 103
+M+E+F QI DLL+L + K + P P +Q + ++ + +
Sbjct 280 KMMKELFHDQIVDLLRLSQENKEAMKKQEVLPTPLCVQGLTDKETFEASHCRASEDAKTT 339
Query 104 WQQPSPHKI------------------WSVEECQKIFLRTFCALMERKQRLLQQQQQQQQ 145
+ P + I WSV+ECQ++F R+F L+ER Q+ +++
Sbjct 340 NKGPKQNAIAPTENTQGEGSGLESQRTWSVQECQEVFERSFLGLLER-----QKTTEREN 394
Query 146 QQQQQQQQGIPFDKDDDLAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNA 205
+++ GIPFDKDDDLAMDFVAAA+ +RM+ FHI K+RW +QA+ G+IIPAIA+TNA
Sbjct 395 AGTGKREAGIPFDKDDDLAMDFVAAAANLRMHNFHIALKSRWFIQAVAGSIIPAIAATNA 454
Query 206 IVAAMQVVEAMHLLSFMEQQKLQKEQQQQQQQPEKTVRDSLARQVWVKPFVTGRRSEEFG 265
+VAA+ QVWVKPFVTG R + G
Sbjct 455 VVAAL--------------------------------------QVWVKPFVTGVRPDAAG 476
Query 266 LLLLPEALEAPRSSCFVCQQITVTVKLASLKQWSLLAFLTKVLQNGLGCKHAF 318
L+LPE ++ PR+SCF+CQQ TVT++LASL W++ F+ ++++ LG H +
Sbjct 477 RLILPEVVDPPRASCFLCQQQTVTIELASLSAWNIETFVERIVKGELGLAHPY 529
> pfa:PFL1790w ubiquitin-activating enzyme, putative
Length=686
Score = 122 bits (306), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 64/237 (27%), Positives = 120/237 (50%), Gaps = 38/237 (16%)
Query 108 SPHKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDF 167
S IW ++C +++++TF L + +++++ + FDKDDD ++F
Sbjct 318 SSQNIWDKKKCIEMYIKTFLKLY------------KYLNINKKEEEYLIFDKDDDECINF 365
Query 168 VAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFME---- 223
+ + S IRM F I K+++++Q+I G IIPAI+STNAIVA++Q + +H++ + E
Sbjct 366 ITSISNIRMLNFCISQKSKFDIQSIAGNIIPAISSTNAIVASLQAFQLIHVIEYFETLKN 425
Query 224 ----------------------QQKLQKEQQQQQQQPEKTVRDSLARQVWVKPFVTGRRS 261
+ + +++ + +R+S A+ VWVK V G +
Sbjct 426 KNNKKNNNNNKKNNNNNNNNNNENNVSYCDEEENKTDHFNIRNSKAKHVWVKSIVNGNKI 485
Query 262 EEFGLLLLPEALEAPRSSCFVCQQITVTVKLASLKQWSLLAFLTKVLQNGLGCKHAF 318
G L+ E LE P +C++CQQ T+ + + + ++ +L F+ + N L + F
Sbjct 486 FSRGNLVNAEPLEIPNPNCYICQQPTIHIYIKNFEKMTLYNFVKDICMNELSFLYPF 542
> tpv:TP01_0127 ubiquitin-protein ligase
Length=543
Score = 117 bits (293), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 78/265 (29%), Positives = 128/265 (48%), Gaps = 32/265 (12%)
Query 49 RVMEEVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQPS 108
++ + +F+ ++K L K+++ W + P+P Q L + Q + + + + +
Sbjct 238 KIFDFLFNSEVKSLQKMEEVWANRDPPKPIKHQFTLKRKASQIDKNSEYDLNSEDTRGRN 297
Query 109 PHKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDFV 168
+ +EE F + A++ + L + F K+D + MDFV
Sbjct 298 KFVVLGMEELLDQFSTSVEAILNNPETL----------------GSLVFSKNDQVCMDFV 341
Query 169 AAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFMEQQKLQ 228
++A+ +RM F I P + W++Q+I GAI+PAIA+TNAIVA+ QVV+ +HLL F++
Sbjct 342 SSAANLRMINFGIKPLSTWDVQSIAGAIVPAIAATNAIVASFQVVQLLHLLKFLKSN--- 398
Query 229 KEQQQQQQQPEKTVRDSLARQVWVKPFVTGRRSEEFGLLLLPEALEAPRSSCFVCQQITV 288
DS ++VW+K V G G L PE LE P C CQQ +
Sbjct 399 ------------NTLDSHCKKVWIKSSVMGSNPLVRGKLSQPELLEPPNPKCTTCQQKSY 446
Query 289 TVKLASLKQWSLLAFLTKVLQNGLG 313
VK+ SL +L F+ VL +G
Sbjct 447 KVKIKSL-DLTLHEFVKSVLSESMG 470
> bbo:BBOV_IV001050 21.m02728; ubiquitin-activating enzyme; K10685
ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=630
Score = 113 bits (282), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 62/161 (38%), Positives = 92/161 (57%), Gaps = 16/161 (9%)
Query 157 FDKDDDLAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAM 216
FDK+D + +DFV++A+ +RM F+IP + W++Q+I G+I PAIA+TNAIVAA QV++ +
Sbjct 407 FDKEDPICVDFVSSAANLRMINFNIPHLSTWDVQSIAGSITPAIAATNAIVAATQVMQLI 466
Query 217 HLLSFMEQQKLQKEQQQQQQQPEKTVRDSLARQ----VWVKPFVTGRRSEEFGLLLLPEA 272
HLL+ + P + SL R VW+K V G G L PE
Sbjct 467 HLLT------------TRHISPGHSADYSLLRDKCKFVWIKSAVAGSAPLTRGALSSPEP 514
Query 273 LEAPRSSCFVCQQITVTVKLASLKQWSLLAFLTKVLQNGLG 313
L+ P C VCQQ + V+L SL W+L +F + + + +G
Sbjct 515 LDEPNPKCAVCQQKVICVELRSLDDWTLESFASTICKMHMG 555
> cpv:cgd2_1460 SUMO-1 activating enzyme subunit 2 ; K10685 ubiquitin-like
1-activating enzyme E1 B [EC:6.3.2.19]
Length=637
Score = 106 bits (265), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 62/209 (29%), Positives = 106/209 (50%), Gaps = 25/209 (11%)
Query 108 SPHKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDF 167
S K++S++E ++F + RK + + + + FDKD+ AMDF
Sbjct 308 SEQKVFSIKENAELFFNS-----ARKIII--------NRMNEIGTASLCFDKDNKDAMDF 354
Query 168 VAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFMEQQKL 227
V+AAS +R FHIP ++RW Q+I G+I+PA+ASTNAIV+ +Q+ + + +L
Sbjct 355 VSAASNLRSYNFHIPLQSRWSCQSIAGSIVPAVASTNAIVSGVQIAQLLLMLKSKLSSLT 414
Query 228 QKEQQQQQQQPEKTVRDSLARQVWVKPFVTGRRSEEFGLLLLPEALEAPRSSCFVCQQIT 287
E + + + VW++ GR ++ PE+LE C +C Q+
Sbjct 415 NPEAGNSDCSSNNKLL-FVNKFVWIRSIPMGR------FIICPESLEKCNPKCLICSQVL 467
Query 288 VTVKLASLKQWSLLAFLTKVLQNGLGCKH 316
V +K+ S +W+L+ F+ G+ C+H
Sbjct 468 VKIKIVSFDKWNLMEFVK-----GIICQH 491
> dre:100333133 ubiquitin-like modifier activating enzyme 2-like;
K10685 ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=642
Score = 95.5 bits (236), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 75/267 (28%), Positives = 133/267 (49%), Gaps = 51/267 (19%)
Query 48 VRVMEEVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQP 107
+++ ++F I LL + K WK K P P L+W ++ Q G Q+Q
Sbjct 253 IKLFNKLFKDDIMYLLTMDKLWKKRKAPLP--------LEW---EEINQLGSQEQVIGS- 300
Query 108 SPHKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDF 167
+++ Q + ++ + L + L+ Q +++ + + +DKDD AMDF
Sbjct 301 ------GLKDQQVLGVQGYAQLFQHSVETLRSQLKEKGDGAE-----LVWDKDDPPAMDF 349
Query 168 VAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFMEQQKL 227
V AAS +RMN F + K+R++++++ G IIPAIA+TNA++A + V+EA+ +L+ +Q
Sbjct 350 VTAASNLRMNVFSMNMKSRFDVKSMAGNIIPAIATTNAVIAGLIVLEALKILNSDFEQ-- 407
Query 228 QKEQQQQQQQPEKTVRDSLARQVWVKPFVTGRRSEEFGLLLLPEALEAPRSSCFVC-QQI 286
R +++ R+ LL+P AL+ P +SC+VC +
Sbjct 408 -------------------CRTIFLNKQPNPRKK-----LLVPCALDPPNASCYVCASKP 443
Query 287 TVTVKLASLKQWSLLAFLTKVLQNGLG 313
VTVKL ++ + + A K+L+ G
Sbjct 444 EVTVKL-NVHKTMVQALQDKILKEKFG 469
> dre:406672 uba2, sae2, sae2b, uble1b, wu:fi17g06, zgc:66354;
ubiquitin-like modifier activating enzyme 2 (EC:6.3.2.-); K10685
ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=640
Score = 95.5 bits (236), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 75/267 (28%), Positives = 133/267 (49%), Gaps = 51/267 (19%)
Query 48 VRVMEEVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQP 107
+++ ++F I LL + K WK K P P L+W ++ Q G Q+Q
Sbjct 253 IKLFNKLFKDDIMYLLTMDKLWKKRKAPLP--------LEW---EEINQLGSQEQVIGS- 300
Query 108 SPHKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDF 167
+++ Q + ++ + L + L+ Q +++ + + +DKDD AMDF
Sbjct 301 ------GLKDQQVLGVQGYAQLFQHSVETLRSQLKEKGDGAE-----LVWDKDDPPAMDF 349
Query 168 VAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFMEQQKL 227
V AAS +RMN F + K+R++++++ G IIPAIA+TNA++A + V+EA+ +L+ +Q
Sbjct 350 VTAASNLRMNVFSMNMKSRFDVKSMAGNIIPAIATTNAVIAGLIVLEALKILNSDFEQ-- 407
Query 228 QKEQQQQQQQPEKTVRDSLARQVWVKPFVTGRRSEEFGLLLLPEALEAPRSSCFVC-QQI 286
R +++ R+ LL+P AL+ P +SC+VC +
Sbjct 408 -------------------CRTIFLNKQPNPRKK-----LLVPCALDPPNASCYVCASKP 443
Query 287 TVTVKLASLKQWSLLAFLTKVLQNGLG 313
VTVKL ++ + + A K+L+ G
Sbjct 444 EVTVKL-NVHKTMVQALQDKILKEKFG 469
> mmu:50995 Uba2, AA986091, Arx, Sae2, UBA1, Ubl1a2, Uble1b; ubiquitin-like
modifier activating enzyme 2 (EC:6.3.2.-); K10685
ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=638
Score = 95.1 bits (235), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 68/263 (25%), Positives = 131/263 (49%), Gaps = 47/263 (17%)
Query 48 VRVMEEVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQP 107
V++ ++F I+ LL + K W+ K P P L W + Q Q + QQ Q
Sbjct 252 VKLFTKLFKDDIRYLLTMDKLWRKRKPPVP--------LDWAEVQSQGEANADQQNEPQ- 302
Query 108 SPHKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDF 167
+++ Q + ++++ +L + L+ ++ + + +DKDD AMDF
Sbjct 303 -----LGLKDQQVLDVKSYASLFSKSIETLRVHLAEKGDGAE-----LIWDKDDPPAMDF 352
Query 168 VAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFMEQQKL 227
V +A+ +RM+ F + K+R++++++ G IIPAIA+TNA++A + V+E + +LS Q
Sbjct 353 VTSAANLRMHIFSMNMKSRFDIKSMAGNIIPAIATTNAVIAGLIVLEGLKILSGKIDQ-- 410
Query 228 QKEQQQQQQQPEKTVRDSLARQVWVKPFVTGRRSEEFGLLLLPEALEAPRSSCFVC-QQI 286
R +++ R+ LL+P AL+ P ++C+VC +
Sbjct 411 -------------------CRTIFLNKQPNPRKK-----LLVPCALDPPNTNCYVCASKP 446
Query 287 TVTVKLASLKQWSLLAFLTKVLQ 309
VTV+L ++ + ++L K+++
Sbjct 447 EVTVRL-NVHKVTVLTLQDKIVK 468
> hsa:10054 UBA2, ARX, FLJ13058, SAE2; ubiquitin-like modifier
activating enzyme 2; K10685 ubiquitin-like 1-activating enzyme
E1 B [EC:6.3.2.19]
Length=640
Score = 94.4 bits (233), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 67/263 (25%), Positives = 130/263 (49%), Gaps = 45/263 (17%)
Query 48 VRVMEEVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQP 107
V++ ++F I+ LL + K W+ K P P L W + Q Q ++ Q +P
Sbjct 252 VKLFTKLFKDDIRYLLTMDKLWRKRKPPVP--------LDWAEVQSQGEETNASDQQNEP 303
Query 108 SPHKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDF 167
+++ Q + ++++ L + L+ ++ + + +DKDD AMDF
Sbjct 304 Q----LGLKDQQVLDVKSYARLFSKSIETLRVHLAEKGDGAE-----LIWDKDDPSAMDF 354
Query 168 VAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFMEQQKL 227
V +A+ +RM+ F + K+R++++++ G IIPAIA+TNA++A + V+E + +LS Q
Sbjct 355 VTSAANLRMHIFSMNMKSRFDIKSMAGNIIPAIATTNAVIAGLIVLEGLKILSGKIDQ-- 412
Query 228 QKEQQQQQQQPEKTVRDSLARQVWVKPFVTGRRSEEFGLLLLPEALEAPRSSCFVC-QQI 286
R +++ R+ LL+P AL+ P +C+VC +
Sbjct 413 -------------------CRTIFLNKQPNPRKK-----LLVPCALDPPNPNCYVCASKP 448
Query 287 TVTVKLASLKQWSLLAFLTKVLQ 309
VTV+L ++ + ++L K+++
Sbjct 449 EVTVRL-NVHKVTVLTLQDKIVK 470
> xla:399235 uba2, MGC84651, Uble1b, sae2, sae2-B, uba2-a, uba2-b,
uble1b-B; ubiquitin-like modifier activating enzyme 2;
K10685 ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=641
Score = 88.6 bits (218), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 62/246 (25%), Positives = 122/246 (49%), Gaps = 46/246 (18%)
Query 48 VRVMEEVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQP 107
+++ ++F IK LL + + W+ K P P L+W ++ + Q +
Sbjct 252 IKLFNKLFRDDIKYLLTMDRLWRKRKPPIP--------LEWASLHNKENCSEIQNE---- 299
Query 108 SPHKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDF 167
+ +++ + + + ++ L + L++Q +++ + + +DKDD AMDF
Sbjct 300 --SSLLGLKDQKVLNVASYAQLFSKSVETLREQLREKGDGAE-----LVWDKDDVPAMDF 352
Query 168 VAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFMEQQKL 227
V AA+ +RM+ F + K++++++++ G IIPAIA+TNA+++ + V+E + +LS +Q
Sbjct 353 VTAAANLRMHIFSMNMKSKFDVKSMAGNIIPAIATTNAVISGLIVLEGLKILSGNTEQ-- 410
Query 228 QKEQQQQQQQPEKTVRDSLARQVWVKPFVTGRRSEEFGLLLLPEALEAPRSSCFVCQ-QI 286
R V++ R+ LL+P +L+ P SC+VC +
Sbjct 411 -------------------CRTVFLNKQPNPRKK-----LLVPCSLDPPNPSCYVCAIKP 446
Query 287 TVTVKL 292
VTVKL
Sbjct 447 EVTVKL 452
> ath:AT2G21470 SAE2; SAE2 (SUMO-ACTIVATING ENZYME 2); SUMO activating
enzyme; K10685 ubiquitin-like 1-activating enzyme E1
B [EC:6.3.2.19]
Length=625
Score = 87.0 bits (214), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 76/320 (23%), Positives = 137/320 (42%), Gaps = 58/320 (18%)
Query 7 ANLLADLKQDLDALCCCSSSSSSSGSGSG--SGSGGGGLAAAGVRVMEEVFSKQIKDLLK 64
A L D QD D ++S+SSS S + G ++ + VF I+ L
Sbjct 190 AKLFGDKNQDNDLNVRSNNSASSSKETEDVFERSEDEDIEQYGRKIYDHVFGSNIEAALS 249
Query 65 LKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQ-----QPS-----PHKIWS 114
++ WK+ + P+P + +L + QQ Q PS P ++W
Sbjct 250 NEETWKNRRRPRP-IYSKDVLPESLTQQNGSTQNCSVTDGDLMVSAMPSLGLKNPQELWG 308
Query 115 VEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDFVAAASII 174
+ + +F+ ++++ + + FDKDD LA++FV AA+ I
Sbjct 309 LTQNSLVFIEALKLFFAKRKKEIGH---------------LTFDKDDQLAVEFVTAAANI 353
Query 175 RMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFMEQQKLQKEQQQQ 234
R +F IP + +E + I G I+ A+A+TNAI+A + V+EA+ +L ++ + K +
Sbjct 354 RAESFGIPLHSLFEAKGIAGNIVHAVATTNAIIAGLIVIEAIKVL----KKDVDKFRMTY 409
Query 235 -QQQPEKTVRDSLARQVWVKPFVTGRRSEEFGLLLLPEALEAPRSSCFVCQQITVTVKLA 293
+ P K LLL+P P +C+VC + + +++
Sbjct 410 CLEHPSKK------------------------LLLMPIEPYEPNPACYVCSETPLVLEIN 445
Query 294 SLKQWSLLAFLTKVLQNGLG 313
+ K L + K+++ LG
Sbjct 446 TRKS-KLRDLVDKIVKTKLG 464
> cel:W02A11.4 uba-2; UBA (human ubiquitin) related family member
(uba-2); K10685 ubiquitin-like 1-activating enzyme E1 B
[EC:6.3.2.19]
Length=582
Score = 85.9 bits (211), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 65/237 (27%), Positives = 104/237 (43%), Gaps = 49/237 (20%)
Query 49 RVMEEVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQPS 108
+V +++F I+ L K++ WK K P P L+++ + Q Q+
Sbjct 249 KVFDKLFLHDIEYLCKMEHLWKQRKRPSP--------LEFHTASSTGGEPQSLCDAQRDD 300
Query 109 PHKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDFV 168
IW++ C K+F + C Q LL+Q + + + + FDKD + M FV
Sbjct 301 T-SIWTLSTCAKVF--STCI-----QELLEQIRAEPDVK-------LAFDKDHAIIMSFV 345
Query 169 AAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFMEQQKLQ 228
AA + IR F IP K++++++A+ G IIPAIASTNAIVA + V EA+ ++
Sbjct 346 AACANIRAKIFGIPMKSQFDIKAMAGNIIPAIASTNAIVAGIIVTEAVRVIE-------- 397
Query 229 KEQQQQQQQPEKTVRDSLARQVWVKPFVTGRRSEEFGLLLLPEALEAPRSSCFVCQQ 285
V + +S G + +A P CFVC +
Sbjct 398 ------------------GSTVICNSSIATTQSNPRGRIFGGDATNPPNPRCFVCSE 436
> sce:YDR390C UBA2, UAL1; Nuclear protein that acts as a heterodimer
with Aos1p to activate Smt3p (SUMO) before its conjugation
to proteins (sumoylation), which may play a role in protein
targeting; essential for viability; K10685 ubiquitin-like
1-activating enzyme E1 B [EC:6.3.2.19]
Length=636
Score = 73.6 bits (179), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 48/180 (26%), Positives = 86/180 (47%), Gaps = 30/180 (16%)
Query 50 VMEEVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQPSP 109
++ ++F + I LL ++ WK+ +P P Q + Q +
Sbjct 253 ILNKLFIQDINKLLAIENLWKTRTKPVP---------------LSDSQINTPTKTAQSAS 297
Query 110 HKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDFVA 169
+ + +++E F+ LM+R + +Q I FDKDD ++FVA
Sbjct 298 NSVGTIQEQISNFINITQKLMDRYPK---------------EQNHIEFDKDDADTLEFVA 342
Query 170 AASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFMEQQKLQK 229
A+ IR + F+IP K+ ++++ I G IIPAIA+TNAIVA + ++ +L+ ++ K
Sbjct 343 TAANIRSHIFNIPMKSVFDIKQIAGNIIPAIATTNAIVAGASSLISLRVLNLLKYAPTTK 402
> dre:406335 uba1, ube1, wu:fa01e08, wu:fb30f01, wu:fi21c11, wu:fj14g11,
zgc:66143; ubiquitin-like modifier activating enzyme
1 (EC:6.3.2.19); K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 61.2 bits (147), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 29/67 (43%), Positives = 42/67 (62%), Gaps = 2/67 (2%)
Query 155 IPFDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQV 212
I F+KDDD MDF+ AAS +R + IPP +R + + I G IIPAIA+T A V +
Sbjct 847 IEFEKDDDTNFHMDFIVAASNLRAENYDIPPADRHKSKLIAGKIIPAIATTTAAVVGLVC 906
Query 213 VEAMHLL 219
+E + ++
Sbjct 907 LELLKIV 913
> xla:398370 uba1-a, MGC68851, a1s9, a1s9t, a1st, amcx1, gxp1,
poc20, smax2, uba1, uba1a, uba1b, ube, ube1, ube1x; ubiquitin-like
modifier activating enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1060
Score = 57.4 bits (137), Expect = 8e-08, Method: Composition-based stats.
Identities = 31/77 (40%), Positives = 45/77 (58%), Gaps = 2/77 (2%)
Query 155 IPFDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQV 212
I F+KDDD MDF+ AAS +R + IPP +R + + I G IIPAIA+T A V +
Sbjct 849 IDFEKDDDTNFHMDFIVAASNLRAENYDIPPADRHKSKLIAGKIIPAIATTTAAVVGLVC 908
Query 213 VEAMHLLSFMEQQKLQK 229
+E ++ + +L K
Sbjct 909 LELYKIIQGHRKLELYK 925
> mmu:74153 Uba7, 1300004C08Rik, Ube1l; ubiquitin-like modifier
activating enzyme 7; K10698 ubiquitin-activating enzyme E1-like
[EC:6.3.2.19]
Length=977
Score = 57.0 bits (136), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 56/101 (55%), Gaps = 12/101 (11%)
Query 132 RKQRLLQQQQQQQQQQQQQQ-----QQGIP-----FDKDDD--LAMDFVAAASIIRMNTF 179
R Q L + Q+Q ++ Q+ ++G P F KDDD +DFV AA+ +R +
Sbjct 738 RPQNLFSAEHGQEQLKELQETLDDWRKGPPLKPVLFVKDDDSNFHVDFVVAATDLRCQNY 797
Query 180 HIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLS 220
I P N ++ IVG IIPAIA++ A+VA + +E ++S
Sbjct 798 GILPVNHARIKQIVGRIIPAIATSTAVVAGLLGLELYKVVS 838
> mmu:22201 Uba1, A1S9, AA989744, Sbx, Ube-1, Ube1x; ubiquitin-like
modifier activating enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 57.0 bits (136), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 38/62 (61%), Gaps = 2/62 (3%)
Query 155 IPFDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQV 212
I F+KDDD MDF+ AAS +R + I P +R + + I G IIPAIA+T A V +
Sbjct 847 IDFEKDDDSNFHMDFIVAASNLRAENYDISPADRHKSKLIAGKIIPAIATTTAAVVGLVC 906
Query 213 VE 214
+E
Sbjct 907 LE 908
> hsa:7317 UBA1, A1S9, A1S9T, A1ST, AMCX1, GXP1, MGC4781, POC20,
SMAX2, UBA1A, UBE1, UBE1X; ubiquitin-like modifier activating
enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating enzyme
E1 [EC:6.3.2.19]
Length=1058
Score = 56.6 bits (135), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 38/62 (61%), Gaps = 2/62 (3%)
Query 155 IPFDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQV 212
I F+KDDD MDF+ AAS +R + IP +R + + I G IIPAIA+T A V +
Sbjct 847 IDFEKDDDSNFHMDFIVAASNLRAENYDIPSADRHKSKLIAGKIIPAIATTTAAVVGLVC 906
Query 213 VE 214
+E
Sbjct 907 LE 908
> xla:379877 uba1-b, MGC52522, a1s9, a1s9t, a1st, amcx1, gxp1,
poc20, smax2, uba1a, ube1, ube1x; ubiquitin-like modifier activating
enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1059
Score = 56.6 bits (135), Expect = 1e-07, Method: Composition-based stats.
Identities = 29/62 (46%), Positives = 39/62 (62%), Gaps = 2/62 (3%)
Query 155 IPFDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQV 212
I F+KDDD MDF+ AAS +R + IPP +R + + I G IIPAIA+T A V +
Sbjct 848 IDFEKDDDTNFHMDFIVAASNLRAENYDIPPADRHKSKLIAGKIIPAIATTTAAVVGLVC 907
Query 213 VE 214
+E
Sbjct 908 LE 909
> mmu:22202 Ube1y1, A1s9Y-1, Sby, Ube-2, Ube1ay, Ube1y, Ube1y-1;
ubiquitin-activating enzyme E1, Chr Y 1; K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 54.7 bits (130), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 38/62 (61%), Gaps = 2/62 (3%)
Query 155 IPFDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQV 212
I F+KDDD MDF+ AAS +R + I P +R + + I G IIPAIA+T + + +
Sbjct 847 IDFEKDDDSNFHMDFIVAASNLRAENYGISPADRHKSKLIAGKIIPAIATTTSAIVGLVC 906
Query 213 VE 214
+E
Sbjct 907 LE 908
> mmu:100040390 ubiquitin-like modifier-activating enzyme 1 Y-like;
K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 54.7 bits (130), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 38/62 (61%), Gaps = 2/62 (3%)
Query 155 IPFDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQV 212
I F+KDDD MDF+ AAS +R + I P +R + + I G IIPAIA+T + + +
Sbjct 847 IDFEKDDDSNFHMDFIVAASNLRAENYGISPADRHKSKLIAGKIIPAIATTTSAIVGLVC 906
Query 213 VE 214
+E
Sbjct 907 LE 908
> tgo:TGME49_090290 ubiquitin-activating enzyme E1, putative ;
K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1091
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 48/76 (63%), Gaps = 3/76 (3%)
Query 155 IPFDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQV 212
+ F+KDDD +D V AAS +R + IP +R + + I G IIPAIA+T A++ +
Sbjct 874 VEFEKDDDTNFHIDLVHAASTLRAMNYKIPCCDRNKTKIIAGRIIPAIATTTAMITGLVS 933
Query 213 VEAMHLLSFMEQQKLQ 228
+E + +++ +Q+KL+
Sbjct 934 LELLKTVTY-KQRKLE 948
> dre:567370 ubiquitin-activating enzyme E1-like; K10699 ubiquitin-activating
enzyme E1-like protein 2 [EC:6.3.2.19]
Length=1060
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 58/122 (47%), Gaps = 14/122 (11%)
Query 157 FDKDDDLA--MDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVE 214
F+KDDD MDFVA+AS +R + I +R + + I G IIPAIA++ A VA + +E
Sbjct 852 FEKDDDTNGHMDFVASASALRARMYAIEAADRLQTKRIAGKIIPAIATSTAAVAGLVSME 911
Query 215 AMHLLS----------FMEQQKLQKEQQQQQQQPEKTVRDSLARQVWVKPFVTGRRSEEF 264
+ + F + Q +RD ++ +W + + GR E+F
Sbjct 912 LIKIAGGYGFELFKNCFFNLAIPVVVLTETAQVKRTQIRDDISFSIWDRWTIFGR--EDF 969
Query 265 GL 266
L
Sbjct 970 TL 971
> hsa:55236 UBA6, E1-L2, FLJ10808, FLJ23367, MOP-4, UBE1L2; ubiquitin-like
modifier activating enzyme 6; K10699 ubiquitin-activating
enzyme E1-like protein 2 [EC:6.3.2.19]
Length=1052
Score = 52.8 bits (125), Expect = 2e-06, Method: Composition-based stats.
Identities = 39/139 (28%), Positives = 68/139 (48%), Gaps = 34/139 (24%)
Query 151 QQQGIPFDKDDDLA--MDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVA 208
Q + F+KDDD +DF+ AAS +R + I P +R++ + I G IIPAIA+T A V+
Sbjct 843 QMAVLSFEKDDDHNGHIDFITAASNLRAKMYSIEPADRFKTKRIAGKIIPAIATTTATVS 902
Query 209 AMQVVE--------------------AMHLLSFMEQQKLQKEQQQQQQQPEKTVRDSLAR 248
+ +E A+ ++ F E +++K + +R+ ++
Sbjct 903 GLVALEMIKVTGGYPFEAYKNCFLNLAIPIVVFTETTEVRKTK----------IRNGISF 952
Query 249 QVWVKPFVTGRRSEEFGLL 267
+W + V G+ E+F LL
Sbjct 953 TIWDRWTVHGK--EDFTLL 969
> mmu:231380 Uba6, 4930542H01, 5730469D23Rik, AU021846, AW124799,
E1-L2, Ube1l2; ubiquitin-like modifier activating enzyme
6; K10699 ubiquitin-activating enzyme E1-like protein 2 [EC:6.3.2.19]
Length=1053
Score = 51.2 bits (121), Expect = 5e-06, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 43/70 (61%), Gaps = 2/70 (2%)
Query 151 QQQGIPFDKDDDLA--MDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVA 208
Q + F+KDDD +DF+ AAS +R + I P +R++ + I G IIPAIA++ A V+
Sbjct 843 QMTVLSFEKDDDRNGHIDFITAASNLRAKMYSIEPADRFKTKRIAGKIIPAIATSTAAVS 902
Query 209 AMQVVEAMHL 218
+ +E + +
Sbjct 903 GLVALEMIKV 912
> ath:AT5G19180 ECR1; ECR1 (E1 C-terminal related 1); NEDD8 activating
enzyme/ protein heterodimerization/ small protein activating
enzyme; K10686 ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=454
Score = 50.1 bits (118), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 45/95 (47%), Gaps = 0/95 (0%)
Query 126 FCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDFVAAASIIRMNTFHIPPKN 185
C L E + + Q + +G FD D+ M +V +I R F IP
Sbjct 214 LCTLAETPRNAAHCIEYAHLIQWETVHRGKTFDPDEPEHMKWVYDEAIRRAELFGIPGVT 273
Query 186 RWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLS 220
Q +V IIPAIASTNAI++A +E + ++S
Sbjct 274 YSLTQGVVKNIIPAIASTNAIISAACALETLKIVS 308
> cpv:cgd4_2300 ubiquitin-activating enzyme E1 (UBA) ; K03178
ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1067
Score = 47.4 bits (111), Expect = 8e-05, Method: Composition-based stats.
Identities = 25/73 (34%), Positives = 42/73 (57%), Gaps = 2/73 (2%)
Query 153 QGIPFDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAM 210
Q I F+KDDD +DF+ + + +R + I +R + + I G IIPA+A+T A++ +
Sbjct 846 QPIEFEKDDDSNFHIDFMNSCANLRARNYSIKECDRHKCKMIAGRIIPAMATTTAMITGL 905
Query 211 QVVEAMHLLSFME 223
EA+ + S E
Sbjct 906 VSFEALKVSSLGE 918
> bbo:BBOV_II007710 18.m06639; ubiquitin-activating enzyme E1;
K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1007
Score = 46.2 bits (108), Expect = 2e-04, Method: Composition-based stats.
Identities = 26/96 (27%), Positives = 53/96 (55%), Gaps = 5/96 (5%)
Query 138 QQQQQQQQQQQQQQQQGIPFDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGA 195
+ + Q+ + + + F+KDDD ++F+ A + +R + I +R + + I G
Sbjct 769 RSKSMQEIMNSRNVFESVEFEKDDDTNYHIEFIWATANLRCQNYDIDQCDRMKAKMISGK 828
Query 196 IIPAIASTNAIVAAMQVVEAMHLLSFMEQQKLQKEQ 231
IIPAIA+T +++A + ++E + + + QKL+ E
Sbjct 829 IIPAIATTTSMIAGLVMLEFVKTICY---QKLKIEH 861
> cel:F11H8.1 rfl-1; ectopic membrane RuFfLes in embryo family
member (rfl-1); K10686 ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=430
Score = 45.8 bits (107), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 43/71 (60%), Gaps = 0/71 (0%)
Query 148 QQQQQQGIPFDKDDDLAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIV 207
+++ +G+ D DD + +++V + +R ++I +R ++ IIPA+ASTNA++
Sbjct 234 EEKPFEGVSLDADDPIHVEWVLERASLRAEKYNIRGVDRRLTSGVLKRIIPAVASTNAVI 293
Query 208 AAMQVVEAMHL 218
AA +EA+ L
Sbjct 294 AASCALEALKL 304
> tpv:TP02_0689 ubiquitin-protein ligase; K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=999
Score = 45.4 bits (106), Expect = 3e-04, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 42/70 (60%), Gaps = 2/70 (2%)
Query 154 GIPFDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQ 211
+ F+KDD+ ++F+ +AS++R + I ++ + + I G IIPAIA+T A++ +
Sbjct 779 AVEFEKDDETNYHIEFIWSASVLRCRNYAIKECDKMKAKLISGKIIPAIATTTAMIGGLV 838
Query 212 VVEAMHLLSF 221
+E + L +
Sbjct 839 TIEFLKALCY 848
> mmu:22200 Uba3, A830034N06Rik, AI256736, AI848246, AW546539,
Ube1c; ubiquitin-like modifier activating enzyme 3; K10686
ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=448
Score = 45.1 bits (105), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 26/94 (27%), Positives = 47/94 (50%), Gaps = 0/94 (0%)
Query 125 TFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDFVAAASIIRMNTFHIPPK 184
T ++ + ++ + Q ++Q G+P D DD + ++ SI R + ++I
Sbjct 224 TIASMPRLPEHCIEYVRMLQWPKEQPFGDGVPLDGDDPEHIQWIFQKSIERASQYNIRGV 283
Query 185 NRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHL 218
Q +V IIPA+ASTNA++AA+ E +
Sbjct 284 TYRLTQGVVKRIIPAVASTNAVIAAVCATEVFKI 317
> xla:734782 uba3, MGC131020, ube1c; ubiquitin-like modifier activating
enzyme 3; K10686 ubiquitin-activating enzyme E1 C
[EC:6.3.2.19]
Length=461
Score = 45.1 bits (105), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 39/72 (54%), Gaps = 0/72 (0%)
Query 147 QQQQQQQGIPFDKDDDLAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAI 206
++Q +G+ D DD ++++ S+ R N F+I Q +V IIPA+ASTNA+
Sbjct 259 KEQPFGEGVQLDGDDPEHIEWIFTNSLERANQFNIRGVTYRLTQGVVKRIIPAVASTNAV 318
Query 207 VAAMQVVEAMHL 218
+AA E +
Sbjct 319 IAAACATEVFKI 330
> hsa:9039 UBA3, DKFZp566J164, MGC22384, UBE1C, hUBA3; ubiquitin-like
modifier activating enzyme 3; K10686 ubiquitin-activating
enzyme E1 C [EC:6.3.2.19]
Length=463
Score = 44.7 bits (104), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 25/94 (26%), Positives = 48/94 (51%), Gaps = 0/94 (0%)
Query 125 TFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDFVAAASIIRMNTFHIPPK 184
T ++ + ++ + Q ++Q +G+P D DD + ++ S+ R + ++I
Sbjct 238 TIASMPRLPEHCIEYVRMLQWPKEQPFGEGVPLDGDDPEHIQWIFQKSLERASQYNIRGV 297
Query 185 NRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHL 218
Q +V IIPA+ASTNA++AA+ E +
Sbjct 298 TYRLTQGVVKRIIPAVASTNAVIAAVCATEVFKI 331
> hsa:7318 UBA7, D8, MGC12713, UBA1B, UBE1L, UBE2; ubiquitin-like
modifier activating enzyme 7; K10698 ubiquitin-activating
enzyme E1-like [EC:6.3.2.19]
Length=1012
Score = 38.9 bits (89), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 35/63 (55%), Gaps = 7/63 (11%)
Query 139 QQQQQQQQQQQQQQQGIP-----FDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQA 191
+QQ++ + + G P F+KDDD +DFV AA+ +R + IPP NR + +
Sbjct 785 EQQKELNKALEVWSVGPPLKPLMFEKDDDSNFHVDFVVAAASLRCQNYGIPPVNRAQSKR 844
Query 192 IVG 194
IVG
Sbjct 845 IVG 847
> dre:100001302 ubiquitin-like modifier-activating enzyme 1-like;
K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1016
Score = 35.8 bits (81), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 24/40 (60%), Gaps = 2/40 (5%)
Query 157 FDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVG 194
F+KDDD MD++ AAS +R + IP +R + + I G
Sbjct 811 FEKDDDSNFHMDYIVAASNLRAENYDIPTADRHKSKLIAG 850
> cel:C47E12.5 uba-1; UBA (human ubiquitin) related family member
(uba-1); K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1113
Score = 32.3 bits (72), Expect = 2.7, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 2/42 (4%)
Query 155 IPFDKDDDLA--MDFVAAASIIRMNTFHIPPKNRWELQAIVG 194
+ F+KDDD M+F+ AAS +R + I P +R + I G
Sbjct 904 VDFEKDDDSNHHMEFITAASNLRAENYDILPADRMRTKQIAG 945
Lambda K H
0.318 0.129 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 13077568040
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40