bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2821_orf2
Length=216
Score E
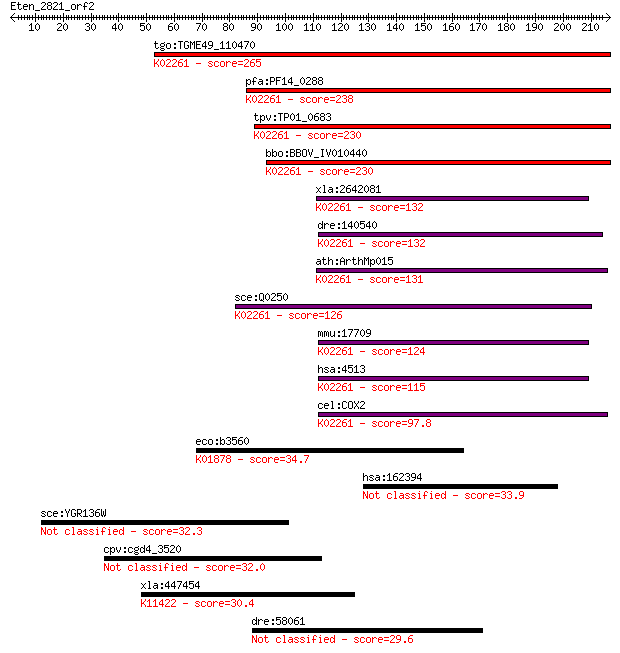
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_110470 cytochrome c oxidase subunit II, putative (E... 265 8e-71
pfa:PF14_0288 cytochrome c oxidase subunit II precursor, putat... 238 9e-63
tpv:TP01_0683 cytochrome c oxidase subunit II precursor; K0226... 230 3e-60
bbo:BBOV_IV010440 23.m05821; cytochrome c oxidase subunit 2 (E... 230 4e-60
xla:2642081 COX2; cytochrome c oxidase subunit II; K02261 cyto... 132 1e-30
dre:140540 COX2, mtco2; cytochrome c oxidase subunit II; K0226... 132 1e-30
ath:ArthMp015 cox2; cytochrome c oxidase subunit 2; K02261 cyt... 131 2e-30
sce:Q0250 COX2, OXI1, OXII; Cox2p (EC:1.9.3.1); K02261 cytochr... 126 5e-29
mmu:17709 COX2; cytochrome c oxidase subunit II (EC:1.9.3.1); ... 124 2e-28
hsa:4513 COX2, COII, MTCO2; cytochrome c oxidase subunit II; K... 115 8e-26
cel:COX2 cytochrome c oxidase subunit II; K02261 cytochrome c ... 97.8 2e-20
eco:b3560 glyQ, cfcA, ECK3548, glyS, JW3531; glycine tRNA synt... 34.7 0.26
hsa:162394 SLFN5, MGC150611, MGC150612, MGC19764; schlafen fam... 33.9 0.41
sce:YGR136W LSB1; Lsb1p 32.3 1.1
cpv:cgd4_3520 hypothetical protein 32.0 1.9
xla:447454 setd1b; SET domain containing 1B (EC:2.1.1.43); K11... 30.4 4.3
dre:58061 hoxa11a; homeo box A11a 29.6 7.4
> tgo:TGME49_110470 cytochrome c oxidase subunit II, putative
(EC:1.9.3.1); K02261 cytochrome c oxidase subunit II [EC:1.9.3.1]
Length=191
Score = 265 bits (677), Expect = 8e-71, Method: Compositional matrix adjust.
Identities = 125/173 (72%), Positives = 142/173 (82%), Gaps = 9/173 (5%)
Query 53 SAAPAAAAAAAAEPE---------VVRPITDAAAAAKSAPAMQNVSAADYPTPAKYLENP 103
AAP AA ++ P +++ + + K +QNV+AADY TP KYL++P
Sbjct 12 GAAPGGAATSSGRPAGCREENSSVLLQSLKANSQQKKGETPLQNVAAADYVTPQKYLDDP 71
Query 104 DLIPTYYTFQSNMVSDEDLLPGMLRNLEVDRRLTLPTRTHIRFLVTATDVIHSWAVPALG 163
D IPTYY FQSNMV+DEDLLPGMLRNLEVD+RLTLPTRTHIRFL+TATDVIHSWAVPALG
Sbjct 72 DKIPTYYVFQSNMVTDEDLLPGMLRNLEVDKRLTLPTRTHIRFLITATDVIHSWAVPALG 131
Query 164 IKADAIPGRLQKVNTFIQREGVFYGQCSELCGALHGFMPIVVEAVSPETYAAH 216
IKADAIPGRLQ++NTFIQREGVFYGQCSELCGALHGFMPIV+EAVSPETYAAH
Sbjct 132 IKADAIPGRLQRINTFIQREGVFYGQCSELCGALHGFMPIVIEAVSPETYAAH 184
> pfa:PF14_0288 cytochrome c oxidase subunit II precursor, putative;
K02261 cytochrome c oxidase subunit II [EC:1.9.3.1]
Length=172
Score = 238 bits (608), Expect = 9e-63, Method: Compositional matrix adjust.
Identities = 106/131 (80%), Positives = 116/131 (88%), Gaps = 0/131 (0%)
Query 86 QNVSAADYPTPAKYLENPDLIPTYYTFQSNMVSDEDLLPGMLRNLEVDRRLTLPTRTHIR 145
+NV A DYP P KYLE+PD IP YY FQSNMV+DEDL PGMLR LEVD+RLTLPTRTHI
Sbjct 35 ENVRAEDYPIPEKYLEDPDKIPKYYVFQSNMVTDEDLQPGMLRQLEVDKRLTLPTRTHIS 94
Query 146 FLVTATDVIHSWAVPALGIKADAIPGRLQKVNTFIQREGVFYGQCSELCGALHGFMPIVV 205
FLVTATDVIHSW++P+LGIKADAIPGRL K+ TFI REGV+YGQCSE+CG LHGFMPIVV
Sbjct 95 FLVTATDVIHSWSIPSLGIKADAIPGRLHKITTFILREGVYYGQCSEMCGTLHGFMPIVV 154
Query 206 EAVSPETYAAH 216
EAVSPE YAAH
Sbjct 155 EAVSPEAYAAH 165
> tpv:TP01_0683 cytochrome c oxidase subunit II precursor; K02261
cytochrome c oxidase subunit II [EC:1.9.3.1]
Length=176
Score = 230 bits (586), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 101/128 (78%), Positives = 114/128 (89%), Gaps = 0/128 (0%)
Query 89 SAADYPTPAKYLENPDLIPTYYTFQSNMVSDEDLLPGMLRNLEVDRRLTLPTRTHIRFLV 148
S YP P KYL+NP+LIP YY+FQSN+V+DEDL PGMLR LEVD+RLTLPTRTHIRFLV
Sbjct 42 SRERYPIPKKYLDNPELIPKYYSFQSNLVTDEDLQPGMLRQLEVDKRLTLPTRTHIRFLV 101
Query 149 TATDVIHSWAVPALGIKADAIPGRLQKVNTFIQREGVFYGQCSELCGALHGFMPIVVEAV 208
TATDV+HSW+VP+LGIK DA+PGRL ++NTFI REGVFYGQCSE+CG LHGFMPIVVEAV
Sbjct 102 TATDVLHSWSVPSLGIKVDAVPGRLTRINTFILREGVFYGQCSEMCGTLHGFMPIVVEAV 161
Query 209 SPETYAAH 216
SPE YA H
Sbjct 162 SPEKYAEH 169
> bbo:BBOV_IV010440 23.m05821; cytochrome c oxidase subunit 2
(EC:1.9.3.1); K02261 cytochrome c oxidase subunit II [EC:1.9.3.1]
Length=168
Score = 230 bits (586), Expect = 4e-60, Method: Compositional matrix adjust.
Identities = 100/124 (80%), Positives = 116/124 (93%), Gaps = 0/124 (0%)
Query 93 YPTPAKYLENPDLIPTYYTFQSNMVSDEDLLPGMLRNLEVDRRLTLPTRTHIRFLVTATD 152
YP P +Y +NPDLIP YY+FQSN+V+DEDL+PGMLR LEVD+RLTLPTRTHIRFL+TATD
Sbjct 37 YPIPEEYEKNPDLIPKYYSFQSNLVTDEDLVPGMLRQLEVDKRLTLPTRTHIRFLITATD 96
Query 153 VIHSWAVPALGIKADAIPGRLQKVNTFIQREGVFYGQCSELCGALHGFMPIVVEAVSPET 212
VIH+W+VP+LGIKADA+PGRL +VNTFI REGV+YGQCSE+CG LHGFMPIVVEAV+PET
Sbjct 97 VIHTWSVPSLGIKADAVPGRLHRVNTFILREGVYYGQCSEMCGTLHGFMPIVVEAVAPET 156
Query 213 YAAH 216
YAAH
Sbjct 157 YAAH 160
> xla:2642081 COX2; cytochrome c oxidase subunit II; K02261 cytochrome
c oxidase subunit II [EC:1.9.3.1]
Length=229
Score = 132 bits (331), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 60/98 (61%), Positives = 71/98 (72%), Gaps = 0/98 (0%)
Query 111 TFQSNMVSDEDLLPGMLRNLEVDRRLTLPTRTHIRFLVTATDVIHSWAVPALGIKADAIP 170
+F S M+ DL PG R LEVD R+ +P + R LVTA DV+HSWAVP+LG+K DAIP
Sbjct 117 SFDSYMIPTNDLTPGQFRLLEVDNRMVVPMESPTRLLVTAEDVLHSWAVPSLGVKTDAIP 176
Query 171 GRLQKVNTFIQREGVFYGQCSELCGALHGFMPIVVEAV 208
GRL + + R GVFYGQCSE+CGA H FMPIVVEAV
Sbjct 177 GRLHQTSFIATRPGVFYGQCSEICGANHSFMPIVVEAV 214
> dre:140540 COX2, mtco2; cytochrome c oxidase subunit II; K02261
cytochrome c oxidase subunit II [EC:1.9.3.1]
Length=230
Score = 132 bits (331), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 61/102 (59%), Positives = 73/102 (71%), Gaps = 0/102 (0%)
Query 112 FQSNMVSDEDLLPGMLRNLEVDRRLTLPTRTHIRFLVTATDVIHSWAVPALGIKADAIPG 171
F S MV +DL PG R LE D R+ +P + IR LV+A DV+HSWAVP+LGIK DA+PG
Sbjct 118 FDSYMVPTQDLTPGGFRLLETDHRMVVPKESPIRILVSAEDVLHSWAVPSLGIKMDAVPG 177
Query 172 RLQKVNTFIQREGVFYGQCSELCGALHGFMPIVVEAVSPETY 213
RL + + R GVFYGQCSE+CGA H FMPIVVEAV E +
Sbjct 178 RLNQTAFIVSRPGVFYGQCSEICGANHSFMPIVVEAVPLEFF 219
> ath:ArthMp015 cox2; cytochrome c oxidase subunit 2; K02261 cytochrome
c oxidase subunit II [EC:1.9.3.1]
Length=260
Score = 131 bits (329), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 58/105 (55%), Positives = 77/105 (73%), Gaps = 0/105 (0%)
Query 111 TFQSNMVSDEDLLPGMLRNLEVDRRLTLPTRTHIRFLVTATDVIHSWAVPALGIKADAIP 170
TF S M+ +EDL G R LEVD R+ +P +TH+R +VT+ DV HSWAVP+ G+K DA+P
Sbjct 143 TFDSYMIPEEDLELGQSRLLEVDNRVVVPAKTHLRIIVTSADVPHSWAVPSSGVKCDAVP 202
Query 171 GRLQKVNTFIQREGVFYGQCSELCGALHGFMPIVVEAVSPETYAA 215
GRL +++ +QREGV+YGQCSE+CG H F IVVEAV + Y +
Sbjct 203 GRLNQISILVQREGVYYGQCSEICGTNHAFTSIVVEAVPRKDYGS 247
> sce:Q0250 COX2, OXI1, OXII; Cox2p (EC:1.9.3.1); K02261 cytochrome
c oxidase subunit II [EC:1.9.3.1]
Length=251
Score = 126 bits (317), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 69/131 (52%), Positives = 84/131 (64%), Gaps = 5/131 (3%)
Query 82 APAMQNVSAADYPTPAKYLENPDLIPT---YYTFQSNMVSDEDLLPGMLRNLEVDRRLTL 138
+PAM + A Y KY E D I F+S ++ DE L G LR L+ D + +
Sbjct 112 SPAM-TIKAIGYQWYWKY-EYSDFINDSGETVEFESYVIPDELLEEGQLRLLDTDTSMVV 169
Query 139 PTRTHIRFLVTATDVIHSWAVPALGIKADAIPGRLQKVNTFIQREGVFYGQCSELCGALH 198
P THIRF+VTA DVIH +A+P+LGIK DA PGRL +V+ IQREGVFYG CSELCG H
Sbjct 170 PVDTHIRFVVTAADVIHDFAIPSLGIKVDATPGRLNQVSALIQREGVFYGACSELCGTGH 229
Query 199 GFMPIVVEAVS 209
MPI +EAVS
Sbjct 230 ANMPIKIEAVS 240
> mmu:17709 COX2; cytochrome c oxidase subunit II (EC:1.9.3.1);
K02261 cytochrome c oxidase subunit II [EC:1.9.3.1]
Length=227
Score = 124 bits (312), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 56/97 (57%), Positives = 69/97 (71%), Gaps = 0/97 (0%)
Query 112 FQSNMVSDEDLLPGMLRNLEVDRRLTLPTRTHIRFLVTATDVIHSWAVPALGIKADAIPG 171
F S M+ DL PG LR LEVD R+ LP IR L+++ DV+HSWAVP+LG+K DAIPG
Sbjct 118 FDSYMIPTNDLKPGELRLLEVDNRVVLPMELPIRMLISSEDVLHSWAVPSLGLKTDAIPG 177
Query 172 RLQKVNTFIQREGVFYGQCSELCGALHGFMPIVVEAV 208
RL + R G+FYGQCSE+CG+ H FMPIV+E V
Sbjct 178 RLNQATVTSNRPGLFYGQCSEICGSNHSFMPIVLEMV 214
> hsa:4513 COX2, COII, MTCO2; cytochrome c oxidase subunit II;
K02261 cytochrome c oxidase subunit II [EC:1.9.3.1]
Length=227
Score = 115 bits (289), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 54/97 (55%), Positives = 67/97 (69%), Gaps = 0/97 (0%)
Query 112 FQSNMVSDEDLLPGMLRNLEVDRRLTLPTRTHIRFLVTATDVIHSWAVPALGIKADAIPG 171
F S M+ L PG LR L+VD R+ LP IR ++T+ DV+HSWAVP LG+K DAIPG
Sbjct 118 FNSYMLPPLFLEPGDLRLLDVDNRVVLPIEAPIRMMITSQDVLHSWAVPTLGLKTDAIPG 177
Query 172 RLQKVNTFIQREGVFYGQCSELCGALHGFMPIVVEAV 208
RL + R GV+YGQCSE+CGA H FMPIV+E +
Sbjct 178 RLNQTTFTATRPGVYYGQCSEICGANHSFMPIVLELI 214
> cel:COX2 cytochrome c oxidase subunit II; K02261 cytochrome
c oxidase subunit II [EC:1.9.3.1]
Length=231
Score = 97.8 bits (242), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 48/104 (46%), Positives = 64/104 (61%), Gaps = 0/104 (0%)
Query 112 FQSNMVSDEDLLPGMLRNLEVDRRLTLPTRTHIRFLVTATDVIHSWAVPALGIKADAIPG 171
F S M S + L G R LEVD R +P T+IRF +T+ DVIH+WA+ +L +K DA+ G
Sbjct 121 FDSYMKSLDQLSLGEPRLLEVDNRCVIPCDTNIRFCITSADVIHAWALNSLSVKLDAMSG 180
Query 172 RLQKVNTFIQREGVFYGQCSELCGALHGFMPIVVEAVSPETYAA 215
L + GVFYGQCSE+CGA H FMPI +E + + +
Sbjct 181 ILSTFSYSFPMVGVFYGQCSEICGANHSFMPIALEVTLLDNFKS 224
> eco:b3560 glyQ, cfcA, ECK3548, glyS, JW3531; glycine tRNA synthetase,
alpha subunit (EC:6.1.1.14); K01878 glycyl-tRNA synthetase
alpha chain [EC:6.1.1.14]
Length=303
Score = 34.7 bits (78), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 33/112 (29%), Positives = 44/112 (39%), Gaps = 27/112 (24%)
Query 68 VVRPITDAAAAAKSAP------------AMQNVSAADYPTPAKYLENPDLIPTYYTFQSN 115
+V+P+ A S P A V + PT +Y ENP+ + YY FQ
Sbjct 27 IVQPLDMEVGAGTSHPMTCLRELGPEPMAAAYVQPSRRPTDGRYGENPNRLQHYYQFQVV 86
Query 116 MVSDED----LLPGMLRNLEVDRRLTLPTRTHIRFLVTATDVIHSWAVPALG 163
+ D L G L+ L +D PT IRF V +W P LG
Sbjct 87 IKPSPDNIQELYLGSLKELGMD-----PTIHDIRF------VEDNWENPTLG 127
> hsa:162394 SLFN5, MGC150611, MGC150612, MGC19764; schlafen family
member 5
Length=891
Score = 33.9 bits (76), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 34/73 (46%), Gaps = 15/73 (20%)
Query 128 RNLEVDRRLTLPTRTHIRFLVTATDVIHSWAVPALGIKADAIPGRLQK-VNTFIQREG-- 184
+ L+ +L LP TH+ F++ +TDV H + RL K V+ F EG
Sbjct 179 KRLQYLEKLNLPESTHVEFVMFSTDVSH------------CVKDRLPKCVSAFANTEGGY 226
Query 185 VFYGQCSELCGAL 197
VF+G E C +
Sbjct 227 VFFGVHDETCQVI 239
> sce:YGR136W LSB1; Lsb1p
Length=241
Score = 32.3 bits (72), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 26/90 (28%), Positives = 47/90 (52%), Gaps = 9/90 (10%)
Query 12 LKMLERMR-CWFSGSKDDRSGLLLPSLTQGGASPALCRSAEVSAAPAAAAAAAAEPEVVR 70
+++LE++ W+ G +++ G+ + + PA RSA +A AA+++ + P V
Sbjct 80 IQVLEKISPDWYRGKSNNKIGIFPANYVK----PAFTRSASPKSAEAASSSTVSRPSVPP 135
Query 71 PITDAAAAAKSAPAMQNVSAADYPTPAKYL 100
P + AA+ + Q VSA Y PA Y+
Sbjct 136 PSYEPAASQYPS---QQVSAP-YAPPAGYM 161
> cpv:cgd4_3520 hypothetical protein
Length=1002
Score = 32.0 bits (71), Expect = 1.9, Method: Composition-based stats.
Identities = 26/99 (26%), Positives = 44/99 (44%), Gaps = 21/99 (21%)
Query 35 PSLTQG----GASPALCRSAEVSAAPAAAAAAAA---------EPEVVRPITDAAAAAKS 81
PSL+Q + PA ++ ++ PA+A+A + +P + + + + + S
Sbjct 627 PSLSQAVVFSTSVPASVPASVPASVPASASAPGSYISQVEPSRDPSLSQTMVSSTSVPAS 686
Query 82 APA-------MQNVSAADYPTPAKY-LENPDLIPTYYTF 112
AP N S A YPTP + + P L PT T+
Sbjct 687 APTPGPAPVPAPNPSLATYPTPVPHPVPGPTLAPTALTY 725
> xla:447454 setd1b; SET domain containing 1B (EC:2.1.1.43); K11422
histone-lysine N-methyltransferase SETD1 [EC:2.1.1.43]
Length=1938
Score = 30.4 bits (67), Expect = 4.3, Method: Composition-based stats.
Identities = 23/77 (29%), Positives = 30/77 (38%), Gaps = 16/77 (20%)
Query 48 RSAEVSAAPAAAAAAAAEPEVVRPITDAAAAAKSAPAMQNVSAADYPTPAKYLENPDLIP 107
R V P A A EP ++RP+T A +S P ++ LE P L
Sbjct 1212 RLEPVPLVPDNAPPAVQEPMIIRPLTPTGAFGESGPVLK-------------LEEPKLQV 1258
Query 108 TYYTFQSNMVSDEDLLP 124
F + DEDL P
Sbjct 1259 NLAHF---VAEDEDLYP 1272
> dre:58061 hoxa11a; homeo box A11a
Length=315
Score = 29.6 bits (65), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 19/84 (22%), Positives = 36/84 (42%), Gaps = 1/84 (1%)
Query 88 VSAADYPTPAKYL-ENPDLIPTYYTFQSNMVSDEDLLPGMLRNLEVDRRLTLPTRTHIRF 146
V AD+ T +L ++P P Y++ SN+ + + R+ +D P R +
Sbjct 23 VPGADFSTLPSFLSQSPSTRPVTYSYASNLPQVQHVREVTFRDYAIDPSTKWPHRGPLAH 82
Query 147 LVTATDVIHSWAVPALGIKADAIP 170
+ D +H +PA+ + P
Sbjct 83 CYPSEDSVHKECLPAVTTVGEMFP 106
Lambda K H
0.318 0.130 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6945028560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40