bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2865_orf2
Length=403
Score E
Sequences producing significant alignments: (Bits) Value
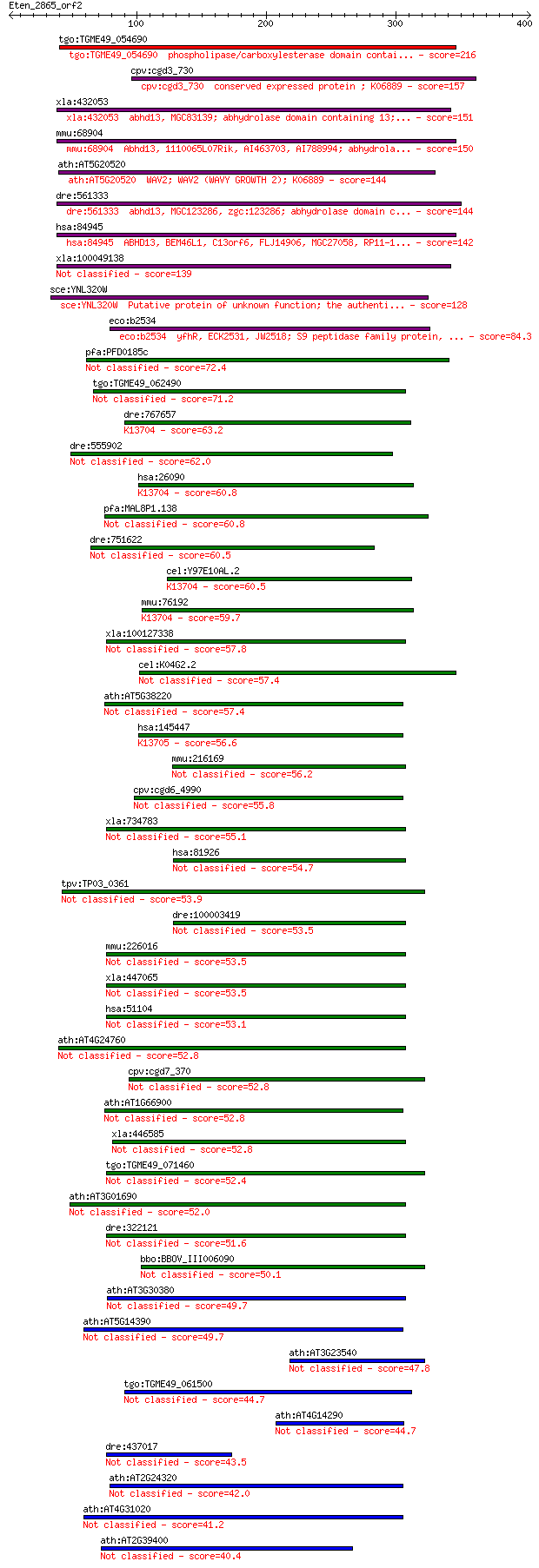
tgo:TGME49_054690 phospholipase/carboxylesterase domain contai... 216 1e-55
cpv:cgd3_730 conserved expressed protein ; K06889 157 5e-38
xla:432053 abhd13, MGC83139; abhydrolase domain containing 13;... 151 5e-36
mmu:68904 Abhd13, 1110065L07Rik, AI463703, AI788994; abhydrola... 150 7e-36
ath:AT5G20520 WAV2; WAV2 (WAVY GROWTH 2); K06889 144 5e-34
dre:561333 abhd13, MGC123286, zgc:123286; abhydrolase domain c... 144 6e-34
hsa:84945 ABHD13, BEM46L1, C13orf6, FLJ14906, MGC27058, RP11-1... 142 3e-33
xla:100049138 hypothetical protein LOC100049138 139 1e-32
sce:YNL320W Putative protein of unknown function; the authenti... 128 5e-29
eco:b2534 yfhR, ECK2531, JW2518; S9 peptidase family protein, ... 84.3 7e-16
pfa:PFD0185c conserved Plasmodium protein, unknown function 72.4 3e-12
tgo:TGME49_062490 hypothetical protein 71.2 6e-12
dre:767657 abhd12, MGC153367, zgc:153367; abhydrolase domain c... 63.2 2e-09
dre:555902 Bem46-like 62.0 4e-09
hsa:26090 ABHD12, ABHD12A, BEM46L2, C20orf22, DKFZp434P106, PH... 60.8 8e-09
pfa:MAL8P1.138 alpha/beta hydrolase, putative 60.8 9e-09
dre:751622 MGC153037, zgc:153037; si:ch211-117n7.7 60.5 1e-08
cel:Y97E10AL.2 hypothetical protein; K13704 abhydrolase domain... 60.5 1e-08
mmu:76192 Abhd12, 1500011G07Rik, 6330583M11Rik, AI431047, AW54... 59.7 2e-08
xla:100127338 hypothetical protein LOC100127338 57.8 8e-08
cel:K04G2.2 hypothetical protein 57.4 1e-07
ath:AT5G38220 hydrolase 57.4 1e-07
hsa:145447 ABHD12B, BEM46L3, C14orf29, MGC129926, MGC129927, c... 56.6 1e-07
mmu:216169 Fam108a, 1700013O15Rik, BC005632, D10Bwg1364e, MGC1... 56.2 2e-07
cpv:cgd6_4990 peptidase of the alpha/beta-hydrolase fold 55.8 3e-07
xla:734783 fam108a1, MGC131027, fam108a2; family with sequence... 55.1 5e-07
hsa:81926 FAM108A1, C19orf27, MGC5244; family with sequence si... 54.7 7e-07
tpv:TP03_0361 hypothetical protein 53.9 1e-06
dre:100003419 si:rp71-61h23.3 53.5 1e-06
mmu:226016 Fam108b, 5730446C15Rik, Cgi67, Fam108b1, MGC40949; ... 53.5 1e-06
xla:447065 fam108b1, MGC83647; abhydrolase domain-containing p... 53.5 2e-06
hsa:51104 FAM108B1, C9orf77, RP11-409O11.2; family with sequen... 53.1 2e-06
ath:AT4G24760 hypothetical protein 52.8 2e-06
cpv:cgd7_370 protein with a conserved N-terminal region 52.8 2e-06
ath:AT1G66900 hypothetical protein 52.8 2e-06
xla:446585 fam108b1, MGC81688; family with sequence similarity... 52.8 2e-06
tgo:TGME49_071460 hypothetical protein 52.4 3e-06
ath:AT3G01690 hypothetical protein 52.0 4e-06
dre:322121 fb50g01, wu:fb50g01; zgc:162293 51.6 6e-06
bbo:BBOV_III006090 17.m07539; hypothetical protein 50.1 1e-05
ath:AT3G30380 hypothetical protein 49.7 2e-05
ath:AT5G14390 hypothetical protein 49.7 2e-05
ath:AT3G23540 hypothetical protein 47.8 7e-05
tgo:TGME49_061500 hypothetical protein 44.7 7e-04
ath:AT4G14290 hypothetical protein 44.7 7e-04
dre:437017 zgc:100937 43.5 0.001
ath:AT2G24320 hypothetical protein 42.0 0.004
ath:AT4G31020 hypothetical protein 41.2 0.008
ath:AT2G39400 hydrolase, alpha/beta fold family protein 40.4 0.013
> tgo:TGME49_054690 phospholipase/carboxylesterase domain containing
protein (EC:3.1.-.-); K06889
Length=497
Score = 216 bits (551), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 123/312 (39%), Positives = 177/312 (56%), Gaps = 40/312 (12%)
Query 40 FQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTENE- 98
FQEKL+F P P K P + SPA G+P+++L+L T DGV++H W +KQ +
Sbjct 47 FQEKLLFYPGVPQGFETPDKNPKGLRSPAERGLPFEELWLRTVDGVKLHCWLIKQKLPQV 106
Query 99 --KAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVIDYRGYGRSEGTPSEAGVYMDAD 155
APT I FHGN G+VG LP LY G+NVL++ YRGYG SEG+P+EAGVY D +
Sbjct 107 AAHAPTLIFFHGNAGNVGFRLPNVELLYKHVGVNVLIVSYRGYGFSEGSPTEAGVYRDGE 166
Query 156 AALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHS 215
AALD ++ +Q N + +AN I F+ G S
Sbjct 167 AALDMLVERQ------------NELHIDANKI----------------------FLFGRS 192
Query 216 MGGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMNNISK 275
+GGAVAIDLA +R +++ G++VENTFTSL + + + R F+ V+ +QR+ M+N K
Sbjct 193 LGGAVAIDLAVQRPHQVRGVIVENTFTSLLDMVWVVFPLLRPFQRTVRILQRLYMDNGEK 252
Query 276 VGSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQKFLVEVPEGDHEETFE--IGGLETK 333
+ L LP+LF+ G +DE + RH +L+E C S K +VP G H +T+E IGG
Sbjct 253 IQRLRLPILFISGQKDELVPTRHMKKLFELCPSPLKEKEDVPLGGHNDTWEWAIGGKSYY 312
Query 334 RALGDFVKLGIK 345
+ F++ ++
Sbjct 313 DRIAAFIQHALQ 324
> cpv:cgd3_730 conserved expressed protein ; K06889
Length=419
Score = 157 bits (398), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 97/269 (36%), Positives = 143/269 (53%), Gaps = 41/269 (15%)
Query 96 ENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVIDYRGYGRSEGTPSEAGVYMDA 154
+ EKAPT + FHGN G++G LPR Y+ G+N+ + YRGYG SEGTPSE G Y+DA
Sbjct 160 QQEKAPTIVFFHGNAGNIGHRLPRFLEFYNLIGVNIFAVSYRGYGDSEGTPSEEGFYLDA 219
Query 155 DAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGH 214
A+L++VL + + + K N +F+ GH
Sbjct 220 KASLEYVLSR-------------------TDVVDK-----------------NMIFLYGH 243
Query 215 SMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMNNIS 274
S+GGAVAIDLA + + G+++ENTFT+++ A Y +F+ F + K IQR+ +++S
Sbjct 244 SIGGAVAIDLASKYN--VTGVILENTFTNIKSVAFRVYPIFKYFGFFFKFIQRLKFDSVS 301
Query 275 KVGSLELPVLFLCGLRDENIKPRHSSRLYEACGS--SQKFLVEVPEGDHEETFEIGGLET 332
K+ ++ P+LF+ G DE I P HS LY GS S K + V G H +T+ GG+E
Sbjct 302 KISRVKSPILFVVGNEDEIIPPTHSVELYMKAGSPKSLKKIYLVSGGSHNDTWIKGGMEF 361
Query 333 KRALGDFVKLGIKEKKKGFATSAKTPLSK 361
L F+ I K S+ ++K
Sbjct 362 YLMLLQFIYNAIDYSKPELEVSSNNLINK 390
> xla:432053 abhd13, MGC83139; abhydrolase domain containing 13;
K06889
Length=336
Score = 151 bits (381), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 98/307 (31%), Positives = 157/307 (51%), Gaps = 51/307 (16%)
Query 38 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTEN 97
F FQ+ L++ +P +P+ + + P GIP++++F+ TKD +R++ L+ T +
Sbjct 58 FKFQDVLLY---FPDQPSSSRLY-----IPMPTGIPHENIFIKTKDNIRLNLILLRYTGD 109
Query 98 EKA--PTFILFHGNYGHVGLTLPRAR-WLYDQGMNVLVIDYRGYGRSEGTPSEAGVYMDA 154
+ PT I FHGN G++G LP A L + +N++++DYRGYG+S+G PSE G+YMD+
Sbjct 110 NSSFSPTIIYFHGNAGNIGHRLPNALLMLVNLKVNLILVDYRGYGKSDGEPSEEGLYMDS 169
Query 155 DAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGH 214
+A LD+V+ + I K + + + G
Sbjct 170 EAVLDYVMTRPD--------------------IDKTK-----------------IILFGR 192
Query 215 SMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMNNIS 274
S+GGAVAI LA + + LV+ENTF S+ A ++V + R+L + +
Sbjct 193 SLGGAVAIHLASENAHRICALVLENTFLSIPHMASTLFSVLPM-RYLPLWCYKNKFLSYR 251
Query 275 KVGSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQKFLVEVPEGDHEETFEIGGLETKR 334
K+ +P LF+ GL D+ I P +LYE S K L P+G H +T++ G T
Sbjct 252 KIVQCRMPSLFISGLSDQLIPPFMMKQLYELSPSRTKRLAIFPDGTHNDTWQCQGYFT-- 309
Query 335 ALGDFVK 341
AL F+K
Sbjct 310 ALEQFIK 316
> mmu:68904 Abhd13, 1110065L07Rik, AI463703, AI788994; abhydrolase
domain containing 13; K06889
Length=337
Score = 150 bits (380), Expect = 7e-36, Method: Compositional matrix adjust.
Identities = 98/311 (31%), Positives = 158/311 (50%), Gaps = 51/311 (16%)
Query 38 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTEN 97
+ FQ+ L++ +P +P+ + + P GIP++++F+ TKDGVR++ ++ T +
Sbjct 58 YKFQDVLLY---FPEQPSSSRLY-----VPMPTGIPHENIFIRTKDGVRLNLILVRYTGD 109
Query 98 EK--APTFILFHGNYGHVGLTLPRAR-WLYDQGMNVLVIDYRGYGRSEGTPSEAGVYMDA 154
PT I FHGN G++G LP A L + +N++++DYRGYG+SEG SE G+Y+D+
Sbjct 110 NSPYCPTIIYFHGNAGNIGHRLPNALLMLVNLRVNLVLVDYRGYGKSEGEASEEGLYLDS 169
Query 155 DAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGH 214
+A LD+V+ + + VF+ G
Sbjct 170 EAVLDYVMTRPDLDKTK-------------------------------------VFLFGR 192
Query 215 SMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMNNIS 274
S+GGAVAI LA + ++ ++VENTF S+ A ++ F + R+L + +
Sbjct 193 SLGGAVAIHLASENSHRISAIMVENTFLSIPHMASTLFSFFPM-RYLPLWCYKNKFLSYR 251
Query 275 KVGSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQKFLVEVPEGDHEETFEIGGLETKR 334
K+ +P LF+ GL D+ I P +LYE S K L P+G H +T++ G T
Sbjct 252 KISQCRMPSLFISGLSDQLIPPVMMKQLYELSPSRTKRLAIFPDGTHNDTWQCQGYFT-- 309
Query 335 ALGDFVKLGIK 345
AL F+K IK
Sbjct 310 ALEQFIKEVIK 320
> ath:AT5G20520 WAV2; WAV2 (WAVY GROWTH 2); K06889
Length=308
Score = 144 bits (363), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 95/305 (31%), Positives = 142/305 (46%), Gaps = 60/305 (19%)
Query 39 SFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTENE 98
+FQEKLV+ P+ P K +P +PA + Y+D++L + DGVR+H WF+K
Sbjct 25 AFQEKLVY---VPVLPGLSKSYPI---TPARLNLIYEDIWLQSSDGVRLHAWFIKMFPEC 78
Query 99 KAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVIDYRGYGRSEGTPSEAGVYMDADAA 157
+ PT + F N G++ L R + + NV ++ YRGYG SEG PS+ G+ DA AA
Sbjct 79 RGPTILFFQENAGNIAHRLEMVRIMIQKLKCNVFMLSYRGYGASEGYPSQQGIIKDAQAA 138
Query 158 LDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSMG 217
LD + G+ +S + V G S+G
Sbjct 139 LDHLSGRTDIDTSR-------------------------------------IVVFGRSLG 161
Query 218 GAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAVFRLFRW-----------LVKAIQ 266
GAV L K ++++ L++ENTFTS+ + A + +W L+ +
Sbjct 162 GAVGAVLTKNNPDKVSALILENTFTSILDMAG---VLLPFLKWFIGGSGTKSLKLLNFVV 218
Query 267 RISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLY--EACGSSQKFLVEVPEGDHEET 324
R I + ++ PVLFL GL+DE + P H LY A + Q VE P G H +T
Sbjct 219 RSPWKTIDAIAEIKQPVLFLSGLQDEMVPPFHMKMLYAKAAARNPQCTFVEFPSGMHMDT 278
Query 325 FEIGG 329
+ GG
Sbjct 279 WLSGG 283
> dre:561333 abhd13, MGC123286, zgc:123286; abhydrolase domain
containing 13; K06889
Length=337
Score = 144 bits (363), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 102/315 (32%), Positives = 160/315 (50%), Gaps = 51/315 (16%)
Query 38 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQT-E 96
+ FQ+ L++ +P +P+ + + P GIP++++++ TKDG+R++ L+ T E
Sbjct 58 YKFQDVLLY---FPDQPSSSRLY-----VPMPTGIPHENVYIRTKDGIRLNLILLRYTGE 109
Query 97 NEK-APTFILFHGNYGHVGLTLPRAR-WLYDQGMNVLVIDYRGYGRSEGTPSEAGVYMDA 154
N APT + FHGN G++G +P A L + NV+++DYRGYG+SEG PSE G+Y DA
Sbjct 110 NPAGAPTILYFHGNAGNIGHRVPNALLMLVNLKANVVLVDYRGYGKSEGDPSEDGLYQDA 169
Query 155 DAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGH 214
+A LD+V+ + I K + V + G
Sbjct 170 EATLDYVMTRPD--------------------IDKTK-----------------VVLFGR 192
Query 215 SMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMNNIS 274
S+GGAVAI LA + +A ++VENTF S+ A ++ F + R+L + +
Sbjct 193 SLGGAVAIRLASCNPHRVAAIMVENTFLSIPHMAATLFSFFPM-RYLPLWCYKNKFLSYR 251
Query 275 KVGSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQKFLVEVPEGDHEETFEIGGLETKR 334
V +P LF+ GL D+ I P +LYE S K L PEG H +T++ G +
Sbjct 252 HVVPCRMPSLFISGLSDQLIPPVMMKQLYELSPSRTKRLAIFPEGTHNDTWQCQGYFS-- 309
Query 335 ALGDFVKLGIKEKKK 349
AL F+K +K +
Sbjct 310 ALEQFMKELLKSHAR 324
> hsa:84945 ABHD13, BEM46L1, C13orf6, FLJ14906, MGC27058, RP11-153I24.2,
bA153I24.2; abhydrolase domain containing 13; K06889
Length=337
Score = 142 bits (358), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 94/311 (30%), Positives = 155/311 (49%), Gaps = 51/311 (16%)
Query 38 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTEN 97
+ FQ+ L++ +P +P+ + + P GIP++++F+ TKDG+R++ ++ T +
Sbjct 58 YKFQDVLLY---FPEQPSSSRLY-----VPMPTGIPHENIFIRTKDGIRLNLILIRYTGD 109
Query 98 EK--APTFILFHGNYGHVGLTLPRARWLYDQGMNVLVI-DYRGYGRSEGTPSEAGVYMDA 154
+PT I FHGN G++G LP A + L++ DYRGYG+SEG SE G+Y+D+
Sbjct 110 NSPYSPTIIYFHGNAGNIGHRLPNALLMLVNLKVNLLLVDYRGYGKSEGEASEEGLYLDS 169
Query 155 DAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGH 214
+A LD+V+ + + +F+ G
Sbjct 170 EAVLDYVMTRPDLDKTK-------------------------------------IFLFGR 192
Query 215 SMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMNNIS 274
S+GGAVAI LA + ++ ++VENTF S+ A ++ F + R+L + +
Sbjct 193 SLGGAVAIHLASENSHRISAIMVENTFLSIPHMASTLFSFFPM-RYLPLWCYKNKFLSYR 251
Query 275 KVGSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQKFLVEVPEGDHEETFEIGGLETKR 334
K+ +P LF+ GL D+ I P +LYE S K L P+G H +T++ G T
Sbjct 252 KISQCRMPSLFISGLSDQLIPPVMMKQLYELSPSRTKRLAIFPDGTHNDTWQCQGYFT-- 309
Query 335 ALGDFVKLGIK 345
AL F+K +K
Sbjct 310 ALEQFIKEVVK 320
> xla:100049138 hypothetical protein LOC100049138
Length=336
Score = 139 bits (351), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 93/307 (30%), Positives = 154/307 (50%), Gaps = 51/307 (16%)
Query 38 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTEN 97
F FQ+ L++ +P +P+ + + P GIP++++F+ TKD +R++ L+ T +
Sbjct 58 FKFQDVLLY---FPDQPSSSRLY-----IPMPTGIPHENIFIKTKDNIRLNLILLRYTGD 109
Query 98 EK--APTFILFHGNYGHVGLTLPRARWLYDQGMNVLVI-DYRGYGRSEGTPSEAGVYMDA 154
+PT + FHGN G++G LP A + L++ DYRGYG+S+G PSE G+Y+D+
Sbjct 110 NSNFSPTIVYFHGNAGNIGHRLPNALLMLVNLKVNLLLVDYRGYGKSDGEPSEEGLYLDS 169
Query 155 DAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGH 214
+A LD+++ + I K + + + G
Sbjct 170 EAVLDYIMTRPD--------------------IDKTK-----------------IILFGR 192
Query 215 SMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMNNIS 274
S+GGAVA+ LA + + LV+ENTF S+ A ++V + R+L + +
Sbjct 193 SLGGAVAVHLASENAHRICALVLENTFLSIPHMASTLFSVLPM-RYLPLWCYKNKFLSYR 251
Query 275 KVGSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQKFLVEVPEGDHEETFEIGGLETKR 334
K+ +P+LF+ GL D+ I P +LYE S K L P+G H +T++ G T
Sbjct 252 KILQCRMPLLFISGLSDQLIPPFMMKQLYELSPSRTKRLAIFPDGTHNDTWQCQGYFT-- 309
Query 335 ALGDFVK 341
AL F+K
Sbjct 310 ALEQFIK 316
> sce:YNL320W Putative protein of unknown function; the authentic,
non-tagged protein is detected in highly purified mitochondria
in high-throughput studies; K06889
Length=284
Score = 128 bits (321), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 93/295 (31%), Positives = 142/295 (48%), Gaps = 53/295 (17%)
Query 33 SIDSEFSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFL 92
S+ + + +Q +LV+ P+ + + + +P S GIPY+ L L T+D +++ W +
Sbjct 20 SVATLYHYQNRLVY-------PSWAQGARNHVDTPDSRGIPYEKLTLITQDHIKLEAWDI 72
Query 93 KQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVIDYRGYGRSEGTPSEAGVY 151
K EN + IL N G++G + Y Q GM+V + YRGYG SEG+PSE G+
Sbjct 73 KN-ENSTSTVLILC-PNAGNIGYFILIIDIFYRQFGMSVFIYSYRGYGNSEGSPSEKGLK 130
Query 152 MDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFV 211
+DAD + S S+ S SK V +
Sbjct 131 LDADCVI------------------------------------SHLSTDSFHSKRKLV-L 153
Query 212 MGHSMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMN 271
G S+GGA A+ +A + + G+++ENTF S+R+ + + + F L I N
Sbjct 154 YGRSLGGANALYIASKFRDLCDGVILENTFLSIRKVIPYIFPLLKRFTLLCHEIW----N 209
Query 272 NISKVGSL--ELPVLFLCGLRDENIKPRHSSRLYEACGSSQKFLVEVPEGDHEET 324
+ +GS E P LFL GL+DE + P H +LYE C SS K + E P G H +T
Sbjct 210 SEGLMGSCSSETPFLFLSGLKDEIVPPFHMRKLYETCPSSNKKIFEFPLGSHNDT 264
> eco:b2534 yfhR, ECK2531, JW2518; S9 peptidase family protein,
function unknown; K06889
Length=284
Score = 84.3 bits (207), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 66/256 (25%), Positives = 112/256 (43%), Gaps = 58/256 (22%)
Query 79 LTTKDGVRIHGWFLKQT---ENEKAPTFILFHGNYGHVGLTLPRARWLYDQGMNVLVIDY 135
T KDG R+ GWF+ + + T I HGN G++ P WL ++ NV + DY
Sbjct 54 FTAKDGTRLQGWFIPSSTGPADNAIATIIHAHGNAGNMSAHWPLVSWLPERNFNVFMFDY 113
Query 136 RGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKS 195
RG+G+S+GTPS+AG+ D +A++ V + ++ Q+
Sbjct 114 RGFGKSKGTPSQAGLLDDTQSAINVVRHRSD--------------------VNPQR---- 149
Query 196 SSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNE-LAGLVVENTFTSLREAAEDT--- 251
+ + G S+GGA +D+ R E + +++++TF S A
Sbjct 150 -------------LVLFGQSIGGANILDVIGRGDREGIRAVILDSTFASYATIANQMIPG 196
Query 252 --YAVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEACGSS 309
Y + + N I+ V +P+L + G D I +HS +LY +
Sbjct 197 SGYLLDESYS---------GENYIASVS--PIPLLLIHGKADHVIPWQHSEKLY-SLAKE 244
Query 310 QKFLVEVPEGDHEETF 325
K L+ +P+G+H + F
Sbjct 245 PKRLILIPDGEHIDAF 260
> pfa:PFD0185c conserved Plasmodium protein, unknown function
Length=734
Score = 72.4 bits (176), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 74/283 (26%), Positives = 117/283 (41%), Gaps = 61/283 (21%)
Query 61 PHIIGSPASYGIPYDDL-FLTTKDGVRIHGWFLKQTENEKAPTFILF-HGNYGHVGLTLP 118
PH P SY +L F+ TK G I G FL N A ILF HGN +G +P
Sbjct 12 PH----PPSYSKNRKNLHFIKTKHGSTICGIFL----NNNAHLTILFSHGNAEDIGDIVP 63
Query 119 R-ARWLYDQGMNVLVIDYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNS 177
+ L G+N+ DY GYG+S G P+E +Y D +AA ++++ +
Sbjct 64 QFESKLKRLGLNMFAYDYSGYGQSTGYPTETHLYNDVEAAYNYLISELN----------- 112
Query 178 NANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVV 237
ISK+ + G S+G A ++ +A +R +L GLV+
Sbjct 113 ---------ISKE-----------------CIIAYGRSLGSAASVHIATKR--DLLGLVL 144
Query 238 ENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPR 297
+ +S+ RL L + NI KV ++ P+LF+ G +D+ +
Sbjct 145 QCPLSSIHRVK------LRLKFTLPYDL----FCNIDKVHLIKCPILFIHGKKDKLLSYH 194
Query 298 HSSRLYEACGSSQKFLVEVPEGDHEETFEIGGLETKRALGDFV 340
+ + + F+ + EG H G + AL F+
Sbjct 195 GTEEMITKTKVNTYFMF-IDEGGHNNLDSCFGNKMTAALVTFL 236
> tgo:TGME49_062490 hypothetical protein
Length=260
Score = 71.2 bits (173), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 65/251 (25%), Positives = 100/251 (39%), Gaps = 71/251 (28%)
Query 66 SPASYGIPYDDLFLTTKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRAR---- 121
SP +Y ++LTT+ RI +F+ + + T I HGN +G+ + +
Sbjct 14 SPPTYECDASFIWLTTRRRQRIPAFFI---DIGASLTIIFSHGNAEDIGMVIEYFKEVSR 70
Query 122 -WLYDQGMNVLVIDYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNAN 180
W N V DY GYG S G PSE GVY +AA +++
Sbjct 71 LW----NCNFFVYDYVGYGHSTGKPSEQGVYDSVEAAFEYL------------------- 107
Query 181 NNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENT 240
Q G +SS + V G S+G + LA R + LAG+++++
Sbjct 108 --------TLQLGLPASS----------IVVYGRSLGTGASCHLASR--HRLAGMILQSG 147
Query 241 FTSLREAAEDTYAVFRLFRWLVKAIQRISM-----NNISKVGSLELPVLFLCGLRDENIK 295
TS+ +T R S+ NI K+G ++ PV + G +DE +
Sbjct 148 LTSIHRVGLNT---------------RFSLPGDMFCNIDKIGRVDCPVFIIHGTKDEIVP 192
Query 296 PRHSSRLYEAC 306
H LY C
Sbjct 193 VHHGMELYNRC 203
> dre:767657 abhd12, MGC153367, zgc:153367; abhydrolase domain
containing 12 (EC:3.1.1.23); K13704 abhydrolase domain-containing
protein 12 [EC:3.1.1.23]
Length=382
Score = 63.2 bits (152), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 61/236 (25%), Positives = 95/236 (40%), Gaps = 57/236 (24%)
Query 90 WFLKQTENEKAPTFILFHGNYGHVG--LTLPRARWLYDQGMNVLVIDYRGYGRSEGTPSE 147
W+ K ++ P + HGN G G + + L G +V+ DYRG+G SEG+PSE
Sbjct 145 WYEKSFQSSH-PVILYLHGNAGTRGGDHRVQLYKVLSSLGYHVVTFDYRGWGDSEGSPSE 203
Query 148 AGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSN 207
G+ DA ++ KQ+ G
Sbjct 204 RGMTSDALFLYQWI---------------------------KQRIGPKP----------- 225
Query 208 FVFVMGHSMGGAVAIDLAKR---RGNELAGLVVENTFTSLREAAED-----TYAVFRLFR 259
+++ GHS+G VA +L +R RG L++E+ FT++RE A+ Y F
Sbjct 226 -LYIWGHSLGTGVATNLVRRLCDRGTPPDALILESPFTNIREEAKSHPFSMVYRYLPGFD 284
Query 260 WLVKAIQRISMNNI-----SKVGSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQ 310
W + IS N+I V + PVL L D + + +LY+ S+
Sbjct 285 WFF--LDAISANDIRFASDENVNHISCPVLILHAEDDTVVPFQLGKKLYDLAAQSK 338
> dre:555902 Bem46-like
Length=344
Score = 62.0 bits (149), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 75/291 (25%), Positives = 114/291 (39%), Gaps = 87/291 (29%)
Query 49 SYPIEPAEIKKFPHIIGSPASYGIPYD-------DLFLTTKDGVRI--------HGWFLK 93
S+ + A I H++ P D +++L ++GVR+ H W
Sbjct 42 SFLVLCATIPTLQHVLSDFVDLSHPLDVGLNHTINVYLKPEEGVRVGVWHTVPEHRWKEA 101
Query 94 QTEN----EKA-----PTFILFHGNYGHVGLTLPR---ARWLYDQGMNVLVIDYRGYGRS 141
Q +N EKA P FI HGN G+ L R A L G +VLV+DYRG+G S
Sbjct 102 QGKNAEWYEKALGDGSPIFIYLHGNGGNRS-ALHRIGVANVLSALGYHVLVMDYRGFGDS 160
Query 142 EGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSS 201
G P+E G+ DA +++ K++ G S
Sbjct 161 TGEPTEPGLTTDALYLYNWI---------------------------KKRSGNS------ 187
Query 202 SSSKSNFVFVMGHSMGGAVAIDLAKR---RGNELAGLVVENTFTSLREAAEDTYAVFRLF 258
V V GHS+G V ++A + G + G+++E S R AA+ F F
Sbjct 188 ------LVCVWGHSIGSGVTTNVAVKLLEEGKKFDGIILEGAMLSGRAAAKQYGHPFSWF 241
Query 259 RWLVKAIQRISMN-------------NISKVGSLELPVLFLCGLRDENIKP 296
W IQ N N+ K+ + P+L L +D+++ P
Sbjct 242 YWKFPYIQFFLFNPLKNNKIVFPLDENLEKIRT---PILILHS-KDDHVSP 288
> hsa:26090 ABHD12, ABHD12A, BEM46L2, C20orf22, DKFZp434P106,
PHARC, dJ965G21.2; abhydrolase domain containing 12 (EC:3.1.1.23);
K13704 abhydrolase domain-containing protein 12 [EC:3.1.1.23]
Length=398
Score = 60.8 bits (146), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 58/225 (25%), Positives = 87/225 (38%), Gaps = 52/225 (23%)
Query 101 PTFILFHGNYGHVG--LTLPRARWLYDQGMNVLVIDYRGYGRSEGTPSEAGVYMDADAAL 158
P + HGN G G + + L G +V+ DYRG+G S GTPSE G+ DA
Sbjct 169 PIILYLHGNAGTRGGDHRVELYKVLSSLGYHVVTFDYRGWGDSVGTPSERGMTYDALHVF 228
Query 159 DFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGG 218
D++ + S N V++ GHS+G
Sbjct 229 DWI---------------------------------------KARSGDNPVYIWGHSLGT 249
Query 219 AVAIDLAKR---RGNELAGLVVENTFTSLREAAED-----TYAVFRLFRW-LVKAIQR-- 267
VA +L +R R L++E+ FT++RE A+ Y F F W + I
Sbjct 250 GVATNLVRRLCERETPPDALILESPFTNIREEAKSHPFSVIYRYFPGFDWFFLDPITSSG 309
Query 268 ISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQKF 312
I N V + P+L L D + + +LY ++ F
Sbjct 310 IKFANDENVKHISCPLLILHAEDDPVVPFQLGRKLYSIAAPARSF 354
> pfa:MAL8P1.138 alpha/beta hydrolase, putative
Length=245
Score = 60.8 bits (146), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 63/262 (24%), Positives = 105/262 (40%), Gaps = 76/262 (29%)
Query 75 DDLFLTTKDGVRIHGWFLKQTENEKAPTFILF-HGNYGHVGLTLPRARWLYDQ------- 126
D +++ T++ ++ F+ N AP ILF HGN +V + LYD
Sbjct 24 DFIYIETENNEKVAAHFI----NRNAPLTILFCHGNGENVYM-------LYDYFYETSKI 72
Query 127 -GMNVLVIDYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNAN 185
+NV + DY GYG S GT SE +Y+ +A D+++ + + +S
Sbjct 73 WNVNVFLYDYLGYGESTGTASEKNMYLSGNAVYDYMVNTLKINPNS-------------- 118
Query 186 AISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTSLR 245
+ + G S+G A+D+A +R ++ GL++++ SL
Sbjct 119 -----------------------IVLYGKSIGSCAAVDIAIKR--KVKGLILQSAILSLL 153
Query 246 EAAEDTYAVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEA 305
T +F S NI ++ + V F+ G D+ + H LYE
Sbjct 154 NICFKTRFIFPF----------DSFCNIKRIKLIPCFVFFIHGTDDKIVPFYHGMCLYEK 203
Query 306 CGSSQKFLVE---VPEGDHEET 324
C KF V V +G H +
Sbjct 204 C----KFKVHPYWVVDGKHNDI 221
> dre:751622 MGC153037, zgc:153037; si:ch211-117n7.7
Length=347
Score = 60.5 bits (145), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 64/247 (25%), Positives = 103/247 (41%), Gaps = 72/247 (29%)
Query 64 IGSPASYGIPYD-DLFLTTKDGVRI--------HGWFLKQTEN----EKA-----PTFIL 105
+ P+ G+ + + +L T++GVR+ H W Q +N EKA P F+
Sbjct 65 LSRPSDLGLNHTINFYLKTEEGVRVGVWHTVPEHRWKEAQGKNVEWYEKALGDGSPIFMY 124
Query 106 FHGNYGH------VGLTLPRARWLYDQGMNVLVIDYRGYGRSEGTPSEAGVYMDADAALD 159
HGN G+ +G+ A L G + LV+DYRG+G S G P+E G+ DA +
Sbjct 125 LHGNTGNRSAPHRIGV----ANILSALGYHALVMDYRGFGDSTGEPTEPGLTTDALYLYN 180
Query 160 FVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGA 219
++ K++ G S + V GHS+G
Sbjct 181 WI---------------------------KKRSGNS------------LLCVWGHSLGSG 201
Query 220 VAIDLAKR---RGNELAGLVVENTFTSLREAAEDTYA-VFRLFRWLVKAIQRISMNNISK 275
V + A + +G + G+++E F S R AA+ + F + W IQ N + K
Sbjct 202 VTTNTAVQLLEQGKKFDGIILEGAFLSGRMAADQVFEHPFTWYYWKFPYIQYFLFNQM-K 260
Query 276 VGSLELP 282
+L+ P
Sbjct 261 NNNLDFP 267
> cel:Y97E10AL.2 hypothetical protein; K13704 abhydrolase domain-containing
protein 12 [EC:3.1.1.23]
Length=345
Score = 60.5 bits (145), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 47/203 (23%), Positives = 86/203 (42%), Gaps = 54/203 (26%)
Query 123 LYDQGMNVLVIDYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNN 182
L D +V+ DYRGYG SEGTP+E G+ D +++
Sbjct 138 LSDCNYHVVCFDYRGYGDSEGTPTEKGIVEDTKTVYEWL--------------------- 176
Query 183 NANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAK---RRGNELAGLVVEN 239
K+ GK+ V V GHSMG V+ L + R GL++E+
Sbjct 177 ------KENCGKTP------------VIVWGHSMGTGVSCKLVQDLSREQQPPCGLILES 218
Query 240 TFTSLREAAEDTYAVFRLFRWL---------VKAIQRI--SMNNISKVGSLELPVLFLCG 288
F +L++A + + +F +F W+ ++ + + +M + ++ + P++ L
Sbjct 219 PFNNLKDAVTN-HPIFTVFSWMNDFMVDHIIIRPLNSVGLTMRSDKRIRLVSCPIIILHA 277
Query 289 LRDENIKPRHSSRLYEACGSSQK 311
D+ + + LYEA +++
Sbjct 278 EDDKILPVKLGRALYEAAKDAER 300
> mmu:76192 Abhd12, 1500011G07Rik, 6330583M11Rik, AI431047, AW547313;
abhydrolase domain containing 12 (EC:3.1.1.23); K13704
abhydrolase domain-containing protein 12 [EC:3.1.1.23]
Length=398
Score = 59.7 bits (143), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 60/223 (26%), Positives = 88/223 (39%), Gaps = 54/223 (24%)
Query 104 ILFHGNYGHVG--LTLPRARWLYDQGMNVLVIDYRGYGRSEGTPSEAGVYMDADAALDFV 161
+ HGN G G + + L G +V+ DYRG+G S GTPSE G+ DA D++
Sbjct 172 LYLHGNAGTRGGDHRVELYKVLSSLGYHVVTFDYRGWGDSVGTPSERGMTYDALHVFDWI 231
Query 162 LGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGAVA 221
+ S N V++ GHS+G VA
Sbjct 232 ---------------------------------------KARSGDNPVYIWGHSLGTGVA 252
Query 222 IDLAKR---RGNELAGLVVENTFTSLREAAED-----TYAVFRLFRW-LVKAIQR--ISM 270
+L +R R L++E+ FT++RE A+ Y F F W + I I
Sbjct 253 TNLVRRLCERETPPDALILESPFTNIREEAKSHPFSVIYRYFPGFDWFFLDPITSSGIKF 312
Query 271 NNISKVGSLELPVLFLCGLRDENIKPRHSSR-LYEACGSSQKF 312
N + + P+L L D+ + P H R LY S+ F
Sbjct 313 ANDENMKHISCPLLILHA-EDDPVVPFHLGRKLYNIAAPSRSF 354
> xla:100127338 hypothetical protein LOC100127338
Length=305
Score = 57.8 bits (138), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 62/237 (26%), Positives = 91/237 (38%), Gaps = 59/237 (24%)
Query 76 DLFLT-TKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQG----MNV 130
D+F+T + G RI +++ + T + HGN +G + D G N+
Sbjct 81 DVFMTKSSRGNRIACMYIRCAPGARF-TLLFSHGNAVDLG---QMTSFYLDLGTRINCNI 136
Query 131 LVIDYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQ 190
DY GYG S G PSE +Y D DAA + +
Sbjct 137 FSYDYSGYGCSSGRPSEKNLYADIDAAWHAL---------------------------RT 169
Query 191 QKGKSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTS-LREAAE 249
+ G S + + + G S+G A+DLA R E A +++ + TS +R
Sbjct 170 RYGISPEN----------ILLYGQSIGTVPAVDLASRY--ECAAVILHSALTSGMRVVLP 217
Query 250 DTYAVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEAC 306
DT + + NI KV + PVL + G DE I H LYE C
Sbjct 218 DTKKTYCF----------DAFPNIEKVSKITSPVLIMHGTEDEVIDFSHGLALYERC 264
> cel:K04G2.2 hypothetical protein
Length=332
Score = 57.4 bits (137), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 63/249 (25%), Positives = 93/249 (37%), Gaps = 57/249 (22%)
Query 102 TFILFHGNYGHVGLTLPRARWLYDQG----MNVLVIDYRGYGRSEGTPSEAGVYMDADAA 157
T + HGN +G +LY G NV DY GYG S G PSE +Y D AA
Sbjct 114 TLLFSHGNAVDLGQ---MTSFLYGLGFHLNCNVFSYDYSGYGCSTGKPSEKNLYADITAA 170
Query 158 LDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSMG 217
+ + + + K++ + + G S+G
Sbjct 171 FELL--------------------KSEFGVPKEK-----------------IILYGQSIG 193
Query 218 GAVAIDLAKRRGNELAGLVVENTFTS-LREAAEDTYAVFRLFRWLVKAIQRISMNNISKV 276
++DLA R +LA LV+ + S +R A T W A +I KV
Sbjct 194 TVPSVDLASRE--DLAALVLHSPLMSGMRVAFPGTTTT-----WCCDAFP-----SIEKV 241
Query 277 GSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQKFLVEVPEGDHEETFEIGGLETKRAL 336
++ P L + G DE I H +YE C +S + L G ++ LE R+
Sbjct 242 PRVKCPTLVIHGTDDEVIDFSHGVSIYERCPTSVEPLWVPGAGHNDVELHAAYLERLRSF 301
Query 337 GDFVKLGIK 345
D I+
Sbjct 302 IDMEASAIR 310
> ath:AT5G38220 hydrolase
Length=336
Score = 57.4 bits (137), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 60/231 (25%), Positives = 91/231 (39%), Gaps = 51/231 (22%)
Query 75 DDLFLTTKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVI 133
D L L T+ G I ++K + T + HGN +G L ++ +N++
Sbjct 43 DVLKLKTRRGNEIVAIYIKHPKANG--TLLYSHGNAADLGQMFELFIELSNRLRLNLMGY 100
Query 134 DYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKG 193
DY GYG+S G SE Y D DAA + K+ G
Sbjct 101 DYSGYGQSTGKASECNTYADIDAAYTCL---------------------------KEHYG 133
Query 194 KSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYA 253
K + + + G S+G IDLA R N L G+V+ + S Y
Sbjct 134 ----------VKDDQLILYGQSVGSGPTIDLASRTPN-LRGVVLHSPILS---GMRVLYP 179
Query 254 VFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYE 304
V R + + + NI K+G++ PVL + G DE + H +L+E
Sbjct 180 VKRTYWFDI-------YKNIDKIGAVTCPVLVIHGTADEVVDCSHGKQLWE 223
> hsa:145447 ABHD12B, BEM46L3, C14orf29, MGC129926, MGC129927,
c14_5314; abhydrolase domain containing 12B; K13705 abhydrolase
domain-containing protein 12B
Length=255
Score = 56.6 bits (135), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 55/218 (25%), Positives = 92/218 (42%), Gaps = 54/218 (24%)
Query 101 PTFILFHGNYGHVGLT--LPRARWLYDQGMNVLVIDYRGYGRSEGTPSEAGVYMDADAAL 158
P + HG+ H + L + L D G +VL +DYRG+G S G P+E G+ DA
Sbjct 33 PIIVYLHGSAEHRAASHRLKLVKVLSDGGFHVLSVDYRGFGDSTGKPTEEGLTTDAICVY 92
Query 159 DFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGG 218
++ +K + G + V + GHS+G
Sbjct 93 EW---------------------------TKARSGITP------------VCLWGHSLGT 113
Query 219 AVAIDLAK---RRGNELAGLVVENTFTSLREAAEDTYAVFRLF-------RWLVKAIQ-- 266
VA + AK +G + +V+E FT++ A+ + Y + +++ R L+ A++
Sbjct 114 GVATNAAKVLEEKGCPVDAIVLEAPFTNMWVASIN-YPLLKIYRNIPGFLRTLMDALRKD 172
Query 267 RISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYE 304
+I N V L P+L L G D + + +LYE
Sbjct 173 KIIFPNDENVKFLSSPLLILHGEDDRTVPLEYGKKLYE 210
> mmu:216169 Fam108a, 1700013O15Rik, BC005632, D10Bwg1364e, MGC11699,
MGC90979; family with sequence similarity 108, member
A
Length=310
Score = 56.2 bits (134), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 52/181 (28%), Positives = 69/181 (38%), Gaps = 50/181 (27%)
Query 127 GMNVLVIDYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANA 186
G N+ DY GYG S G PSE +Y D DAA +
Sbjct 139 GCNIFSYDYSGYGISSGRPSEKNLYADIDAAWQAL------------------------- 173
Query 187 ISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTS-LR 245
+ + G S S + + G S+G +DLA R E A +V+ + TS +R
Sbjct 174 --RTRYGISPDS----------IILYGQSIGTVPTVDLASR--YECAAVVLHSPLTSGMR 219
Query 246 EAAEDTYAVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEA 305
A DT + + NI KV + PVL + G DE I H LYE
Sbjct 220 VAFPDTKKTYCF----------DAFPNIEKVSKITSPVLIIHGTEDEVIDFSHGLALYER 269
Query 306 C 306
C
Sbjct 270 C 270
> cpv:cgd6_4990 peptidase of the alpha/beta-hydrolase fold
Length=383
Score = 55.8 bits (133), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 57/225 (25%), Positives = 86/225 (38%), Gaps = 56/225 (24%)
Query 98 EKAPTFILFHGNYGHVGLTLPRARWLYDQGM----NVLVIDYRGYGRSEGTPSEAGVYMD 153
EK P FI HGN +G LP W + + +VL DYR YG S+G P+E G+Y D
Sbjct 153 EKIPVFIFSHGNATDIGSMLP---WFVNLSLKLNAHVLAYDYRSYGLSKGKPTERGIYAD 209
Query 154 ADAALD-------------FVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSS 200
A + F+LG+ S+ + + N + +
Sbjct 210 IKAVYEYARDELNFPTDRIFLLGQSIGSAPTVHLARKLRKKLKKN-----------TGAG 258
Query 201 SSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTS-LREAAEDTYAVFRLFR 259
++S KSN ID R G L G+++++ S L Y
Sbjct 259 TTSDKSN--------------ID-CNRSGLPLGGIIIQSGIASGLNALLAPDYK------ 297
Query 260 WLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYE 304
K I N + + P+L L G D+ I +S +L+E
Sbjct 298 ---KDIPCDVFPNYRNIRKVPFPILILHGTNDQVIHISNSKKLFE 339
> xla:734783 fam108a1, MGC131027, fam108a2; family with sequence
similarity 108, member A1
Length=305
Score = 55.1 bits (131), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 61/237 (25%), Positives = 91/237 (38%), Gaps = 59/237 (24%)
Query 76 DLFLT-TKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQG----MNV 130
++F+T + G RI +++ + T + HGN +G + D G N+
Sbjct 81 EVFMTKSSRGNRIACMYVRCAPGARF-TLLFSHGNAVDLG---QMTSFYLDLGTRINCNI 136
Query 131 LVIDYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQ 190
DY GYG S G PSE +Y D DAA + +
Sbjct 137 FSYDYSGYGCSSGRPSEKNLYADIDAAWHAL---------------------------RT 169
Query 191 QKGKSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTS-LREAAE 249
+ G S + + + G S+G A+DLA R E A +++ + TS +R
Sbjct 170 RYGISPEN----------ILLYGQSIGTVPAVDLASRY--ECAAVILHSAMTSGMRVVLP 217
Query 250 DTYAVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEAC 306
DT + + NI KV + PVL + G DE I H LYE C
Sbjct 218 DTKKTYCF----------DAFPNIEKVSKITSPVLIMHGTEDEVIDFSHGLALYERC 264
> hsa:81926 FAM108A1, C19orf27, MGC5244; family with sequence
similarity 108, member A1
Length=310
Score = 54.7 bits (130), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 51/180 (28%), Positives = 68/180 (37%), Gaps = 50/180 (27%)
Query 128 MNVLVIDYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAI 187
N+ DY GYG S G PSE +Y D DAA +
Sbjct 140 CNIFSYDYSGYGASSGRPSERNLYADIDAAWQAL-------------------------- 173
Query 188 SKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTS-LRE 246
+ + G S S + + G S+G +DLA R E A +V+ + TS +R
Sbjct 174 -RTRYGISPDS----------IILYGQSIGTVPTVDLASR--YECAAVVLHSPLTSGMRV 220
Query 247 AAEDTYAVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEAC 306
A DT + + NI KV + PVL + G DE I H LYE C
Sbjct 221 AFPDTKKTYCF----------DAFPNIEKVSKITSPVLIIHGTEDEVIDFSHGLALYERC 270
> tpv:TP03_0361 hypothetical protein
Length=315
Score = 53.9 bits (128), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 64/285 (22%), Positives = 104/285 (36%), Gaps = 79/285 (27%)
Query 42 EKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTENEKAP 101
+ L+F P PI P+ + PH+ P T DG I +F+K +
Sbjct 6 DSLIFRP--PIPPSYSRDDPHLHLIP-------------TPDGNTIASYFIKHKFAKF-- 48
Query 102 TFILFHGNYGHVG-----LTLPRARWLYDQGMNVLVIDYRGYGRSEGTPSEAGVYMDADA 156
T I H N +G L +W N+ + DY GYG S G SE +Y AD
Sbjct 49 TIIFSHANAEDIGNVFGNLIKRLTKW----NCNLFIYDYPGYGLSSGVCSEENMYNCADL 104
Query 157 ALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSM 216
+ ++++ + +S + + G S+
Sbjct 105 SYNYLINTLKVNSGN-------------------------------------IIAYGRSL 127
Query 217 GGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMNNISKV 276
G AI L + +L G+++++ F S+ + F F NN KV
Sbjct 128 GCTCAIYLGVKY--KLLGVILQSPFLSIYRIKVPCFLPFDRF------------NNYDKV 173
Query 277 GSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQKFLVEVPEGDH 321
L P L + G D+ I +HS +L + + V+ G+H
Sbjct 174 KDLNCPALVIHGDSDDIIPVQHSIQLIKRIPDVYYYFVKT--GNH 216
> dre:100003419 si:rp71-61h23.3
Length=324
Score = 53.5 bits (127), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 51/180 (28%), Positives = 67/180 (37%), Gaps = 50/180 (27%)
Query 128 MNVLVIDYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAI 187
N+ DY GYG S G PSE +Y D DAA Q+ S S N
Sbjct 154 CNIFSYDYSGYGVSTGKPSEKNLYADIDAAW-------QALRSRYGISPEN--------- 197
Query 188 SKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTS-LRE 246
+ + G S+G +DLA R E A +++ + TS +R
Sbjct 198 ---------------------IILYGQSIGTVPTVDLASRY--ECAAVILHSPLTSGMRV 234
Query 247 AAEDTYAVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEAC 306
A DT + + NI KV + PVL + G DE I H LYE C
Sbjct 235 AFPDTKKTYCF----------DAFPNIEKVSKITSPVLIIHGTEDEVIDFSHGLALYERC 284
> mmu:226016 Fam108b, 5730446C15Rik, Cgi67, Fam108b1, MGC40949;
family with sequence similarity 108, member B
Length=288
Score = 53.5 bits (127), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 61/234 (26%), Positives = 89/234 (38%), Gaps = 53/234 (22%)
Query 76 DLFLT-TKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVI 133
+ F+T T G RI F++ + N K T + HGN +G L + N+
Sbjct 67 ECFMTRTSKGNRIACMFVRCSPNAKY-TLLFSHGNAVDLGQMSSFYIGLGSRINCNIFSY 125
Query 134 DYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKG 193
DY GYG S G P+E +Y D +AA L + N
Sbjct 126 DYSGYGASSGKPTEKNLYADVEAAW---LALRTRYGIRPEN------------------- 163
Query 194 KSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTS-LREAAEDTY 252
V + G S+G ++DLA R E A +++ + TS +R A DT
Sbjct 164 ---------------VIIYGQSIGTVPSVDLAARY--ESAAVILHSPLTSGMRVAFPDTK 206
Query 253 AVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEAC 306
+ + NI K+ + PVL + G DE I H L+E C
Sbjct 207 KTYCF----------DAFPNIDKISKITSPVLIIHGTEDEVIDFSHGLALFERC 250
> xla:447065 fam108b1, MGC83647; abhydrolase domain-containing
protein FAM108B1
Length=288
Score = 53.5 bits (127), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 61/234 (26%), Positives = 90/234 (38%), Gaps = 53/234 (22%)
Query 76 DLFLT-TKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVI 133
+ F+T T G RI F++ + K T + HGN +G L + N+
Sbjct 67 ECFMTRTSRGNRIACMFVRCCPSAKY-TLLFSHGNAVDLGQMSSFYIGLGSRINCNIFSY 125
Query 134 DYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKG 193
DY GYG S G PSE +Y D DAA I+ + +
Sbjct 126 DYSGYGSSSGKPSEKNLYADIDAAW----------------------------IALRTR- 156
Query 194 KSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTS-LREAAEDTY 252
+ V + G S+G ++DLA R E A +++ + TS +R A DT
Sbjct 157 --------YGIRPEHVIIYGQSIGTVPSVDLAARY--ESAAVILHSPLTSGMRVAFPDTK 206
Query 253 AVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEAC 306
+ + NI K+ + PVL + G DE I H L+E C
Sbjct 207 KTYCF----------DAFPNIDKISKITSPVLIIHGTEDEVIDFSHGLALFERC 250
> hsa:51104 FAM108B1, C9orf77, RP11-409O11.2; family with sequence
similarity 108, member B1
Length=288
Score = 53.1 bits (126), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 61/234 (26%), Positives = 89/234 (38%), Gaps = 53/234 (22%)
Query 76 DLFLT-TKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVI 133
+ F+T T G RI F++ + N K T + HGN +G L + N+
Sbjct 67 ECFMTRTSKGNRIACMFVRCSPNAKY-TLLFSHGNAVDLGQMSSFYIGLGSRINCNIFSY 125
Query 134 DYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKG 193
DY GYG S G P+E +Y D +AA L + N
Sbjct 126 DYSGYGASSGKPTEKNLYADIEAAW---LALRTRYGIRPEN------------------- 163
Query 194 KSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTS-LREAAEDTY 252
V + G S+G ++DLA R E A +++ + TS +R A DT
Sbjct 164 ---------------VIIYGQSIGTVPSVDLAARY--ESAAVILHSPLTSGMRVAFPDTK 206
Query 253 AVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEAC 306
+ + NI K+ + PVL + G DE I H L+E C
Sbjct 207 KTYCF----------DAFPNIDKISKITSPVLIIHGTEDEVIDFSHGLALFERC 250
> ath:AT4G24760 hypothetical protein
Length=365
Score = 52.8 bits (125), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 66/274 (24%), Positives = 101/274 (36%), Gaps = 59/274 (21%)
Query 39 SFQEKLVF----DPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQ 94
S KL F PSY + E + + SP + D L L T+ G I +++
Sbjct 7 SMAAKLAFFPPNPPSYKLVRDETTEL--FLMSPFPHRENVDILRLPTRRGTEIVAMYIRY 64
Query 95 TENEKAPTFILF-HGNYGHVGLTLPRARWL-YDQGMNVLVIDYRGYGRSEGTPSEAGVYM 152
A T +L+ HGN +G L +N++ DY GYG+S G P+E Y
Sbjct 65 P---MAVTTLLYSHGNAADIGQMYELFIELSIHLRVNLMGYDYSGYGQSSGKPTEQNTYA 121
Query 153 DADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVM 212
D +AA + N A N + +
Sbjct 122 DIEAAYKCL------------EENYGAKQEN-------------------------IILY 144
Query 213 GHSMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMNN 272
G S+G +DLA R A ++ + LR Y V R + + + N
Sbjct 145 GQSVGSGPTVDLAARLPRLRASILHSPILSGLRV----MYPVKRTYWFDI-------YKN 193
Query 273 ISKVGSLELPVLFLCGLRDENIKPRHSSRLYEAC 306
I K+ + PVL + G D+ + H +L+E C
Sbjct 194 IDKITLVRCPVLVIHGTADDVVDFSHGKQLWELC 227
> cpv:cgd7_370 protein with a conserved N-terminal region
Length=611
Score = 52.8 bits (125), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 52/248 (20%), Positives = 97/248 (39%), Gaps = 61/248 (24%)
Query 94 QTENEKAPTFILFHGNYGHVGLTLPRARWLYDQGMNVLVIDYRGYGRSEGTPSEAGVYM- 152
+ ++E P I HGN LP L G+ V+ +D G G S+G G +
Sbjct 70 ERQSESLPCVIYLHGNCSSRREALPYIPLLLPIGITVMAVDLSGSGLSDGDYISLGYHEK 129
Query 153 -DADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFV 211
D ++++ ++ SS V V
Sbjct 130 DDLSVLVEYLRNSKRCSS---------------------------------------VGV 150
Query 212 MGHSMGGAVAIDLAKRRGNE--LAGLVVENTFTSLREAAEDTY-------------AVFR 256
G SMG A A+ + + L G+V++++F SLR+ + +
Sbjct 151 WGRSMGAATALMYSGVDKGDGFLKGIVIDSSFCSLRQLCHELVHHYIPLLPNFLVDSALS 210
Query 257 LFRWLVKAIQRISMNNIS---KVGSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQKFL 313
+ + ++++++I+ VG ++P LF+ G D + P HS L++ + +K L
Sbjct 211 FIKSTINDKAKVNIDDIAPIKSVGQCKVPALFISGTNDTLVNPNHSKTLHDNY-AGEKML 269
Query 314 VEVPEGDH 321
+ +P G+H
Sbjct 270 MIIP-GNH 276
> ath:AT1G66900 hypothetical protein
Length=272
Score = 52.8 bits (125), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 60/234 (25%), Positives = 95/234 (40%), Gaps = 57/234 (24%)
Query 75 DDLFLTTKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVI 133
D L L T+ G I ++K ++ T + HGN +G L ++ +N++
Sbjct 46 DILKLRTRCGNEIVAVYVKHSKANG--TLLYSHGNAADLGQMFELFVELSNRLRVNLMGY 103
Query 134 DYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKG 193
DY GYG+S G SE Y D +A+ + K++ G
Sbjct 104 DYSGYGQSTGQASECNTYADIEASYKCL---------------------------KEKYG 136
Query 194 KSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTS---LREAAED 250
K + + V G S+G +DLA R N L G+V++ S + +
Sbjct 137 ----------VKDDQLIVYGQSVGSGPTVDLASRTPN-LRGVVLQCPILSGMRVLYPVKC 185
Query 251 TYAVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYE 304
TY F +++ NI K+GS+ PVL + G DE + H RL+E
Sbjct 186 TYW-FDIYK------------NIDKIGSVTCPVLVIHGTADEVVDWSHGKRLWE 226
> xla:446585 fam108b1, MGC81688; family with sequence similarity
108, member B1
Length=288
Score = 52.8 bits (125), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 59/228 (25%), Positives = 87/228 (38%), Gaps = 52/228 (22%)
Query 81 TKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVIDYRGYG 139
T G RI F++ + + K T + HGN +G L + N+ DY GYG
Sbjct 73 TSRGNRIACMFVRCSPSAKY-TLLFSHGNAVDLGQMSSFYIGLGSRINCNIFSYDYSGYG 131
Query 140 RSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSS 199
S G PSE +Y D DAA I+ + +
Sbjct 132 SSSGKPSEKNLYADIDAAW----------------------------IALRTR------- 156
Query 200 SSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTS-LREAAEDTYAVFRLF 258
+ V + G S+G ++DLA R E A +++ + TS +R A DT +
Sbjct 157 --YGVRPEHVIIYGQSIGTVPSVDLAARY--ESAAVILHSPLTSGMRVAFPDTKKTYCF- 211
Query 259 RWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEAC 306
+ NI K+ + PVL + G DE I H L+E C
Sbjct 212 ---------DAFPNIDKISKITSPVLIIHGTEDEVIDFSHGLALFERC 250
> tgo:TGME49_071460 hypothetical protein
Length=657
Score = 52.4 bits (124), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 60/267 (22%), Positives = 102/267 (38%), Gaps = 61/267 (22%)
Query 76 DLFLTTKDGVRIHGWFLKQTE----NEKAPTFILFHGNYGHVGLTLPRARWLYDQGMNVL 131
DL L + R+ + TE EK P + HGN L L Q + V
Sbjct 48 DLELANRRNQRLQCSHYEPTEPFRPQEKLPCVVYLHGNCSSRVEALGTLPVLLPQDITVF 107
Query 132 VIDYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQ 191
D+ G G+S+G G + D LD V+ +++ S+
Sbjct 108 AFDFAGSGKSDGEYVSLGWWERED--LDVVIEHLRATGRVST------------------ 147
Query 192 KGKSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDT 251
+ + G SMG A+ L R + G+V+++ F SLR AE+
Sbjct 148 -----------------IGLWGRSMGAVTAL-LHADRDPSIGGMVLDSPFASLRRLAEEL 189
Query 252 YAV---FRLFRWLVKAIQRI-----------SMNNISKVGSLE---LPVLFLCGLRDENI 294
V ++L R+++ ++ + +NN++ + +E +P +F+ D I
Sbjct 190 AGVVVSWKLPRFVLNSLLAMVRTTIINKAAFDINNLAPIDHVEHTFIPAMFVVANNDTFI 249
Query 295 KPRHSSRLYEACGSSQKFLVEVPEGDH 321
P H L++ + L EGDH
Sbjct 250 LPSHGEELHDKYAGDRNILRV--EGDH 274
> ath:AT3G01690 hypothetical protein
Length=361
Score = 52.0 bits (123), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 59/260 (22%), Positives = 97/260 (37%), Gaps = 53/260 (20%)
Query 48 PSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTENEKAPTFILFH 107
PSY + E+ ++ SP + + + L T+ G I G +++ T + H
Sbjct 20 PSYKVVTDELTGL--LLLSPFPHRENVEIVKLRTRRGTEIVGMYVRHPM--ATSTLLYSH 75
Query 108 GNYGHVGLTLPRARWL-YDQGMNVLVIDYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQ 166
GN +G L +N++ DY GYG+S G PSE Y D +A +
Sbjct 76 GNAADLGQMYELFIELSIHLKVNLMGYDYSGYGQSTGKPSEHNTYADIEAVYKCL----- 130
Query 167 SSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAK 226
+ SK V + G S+G +DLA
Sbjct 131 --------------------------------EETFGSKQEGVILYGQSVGSGPTLDLAS 158
Query 227 RRGNELAGLVVENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMNNISKVGSLELPVLFL 286
R A ++ + LR Y+V + + + + NI K+ ++ PVL +
Sbjct 159 RLPQLRAVVLHSPILSGLRV----MYSVKKTYWFDI-------YKNIDKIPYVDCPVLII 207
Query 287 CGLRDENIKPRHSSRLYEAC 306
G DE + H +L+E C
Sbjct 208 HGTSDEVVDCSHGKQLWELC 227
> dre:322121 fb50g01, wu:fb50g01; zgc:162293
Length=336
Score = 51.6 bits (122), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 60/234 (25%), Positives = 91/234 (38%), Gaps = 53/234 (22%)
Query 76 DLFLT-TKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVI 133
++FLT + G R+ +++ + + T + HGN +G L + N+
Sbjct 115 EVFLTHSSRGNRVGCMYIRCAPSARY-TVLFSHGNAVDLGQMSSFYIGLGTRINCNIFSY 173
Query 134 DYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKG 193
DY GYG S G PSE +Y D DAA + + + G
Sbjct 174 DYSGYGVSTGKPSEKNLYADIDAAWHAL---------------------------RSRYG 206
Query 194 KSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTS-LREAAEDTY 252
S + + + G S+G +DLA R E A +V+ + TS +R A DT
Sbjct 207 ISPEN----------IILYGQSIGTVPTVDLASRY--ECAAVVLHSPLTSGMRVAFPDTK 254
Query 253 AVFRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEAC 306
+ + NI KV + PVL + G DE I H L+E C
Sbjct 255 KTYCF----------DAFPNIEKVSKITSPVLIIHGTEDEVIDFSHGLALFERC 298
> bbo:BBOV_III006090 17.m07539; hypothetical protein
Length=420
Score = 50.1 bits (118), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 53/225 (23%), Positives = 89/225 (39%), Gaps = 49/225 (21%)
Query 103 FILFHGNYGHVGLTLPRARWL-YDQGMNVLVIDYRGYGRSEGTPSEAGVYMDAD--AALD 159
I HGN +G P R + ++ +N+L ++Y GYG S G ++ A +
Sbjct 64 LIYLHGNSCDIGQVKPELRLVAHELNVNILAVEYPGYGVSPEVSVATGELINCRVRATFN 123
Query 160 FVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGA 219
F+L S N +S + G S+G
Sbjct 124 FLL-------SLGVNPHS-------------------------------IIFFGRSIGTG 145
Query 220 VAIDLA---KRRGNELAGLVVENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMNNISKV 276
A LA K+RG + G+++++ + S+ E+ +A L WLV + +++ +
Sbjct 146 PAAALAAEFKKRGIQCGGVILQSPYISIHRIIEEYFA---LGTWLVNNFWD-TEKSLANM 201
Query 277 GSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQKFLVEVPEGDH 321
G + P+L + GL DE + H LYE+ S K P H
Sbjct 202 GP-QTPLLIIHGLADEIVPVYHGQTLYESYKSDIKMADFQPNSKH 245
> ath:AT3G30380 hypothetical protein
Length=377
Score = 49.7 bits (117), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 56/232 (24%), Positives = 87/232 (37%), Gaps = 53/232 (22%)
Query 77 LFLTTKDGVRIHGWFLKQTENEKAPTFILF-HGNYGHVGLTLPRARWL-YDQGMNVLVID 134
L L TK G ++ ++K N A +L+ HGN +G L +N++ D
Sbjct 46 LKLKTKRGNQVVAAYIK---NPTASLTLLYSHGNAADLGQMFELFSELSLHLRVNLIGYD 102
Query 135 YRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGK 194
Y GYGRS G PSE Y D +A + K
Sbjct 103 YSGYGRSSGKPSEQNTYSDIEAVYRCLEEKY----------------------------- 133
Query 195 SSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAV 254
K V + G S+G ++LA R N L +V+ + S Y V
Sbjct 134 --------GVKEQDVILYGQSVGSGPTLELASRLPN-LRAVVLHSAIAS---GLRVMYPV 181
Query 255 FRLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEAC 306
R + + + N+ K+ ++ PVL + G D+ + H +L+E C
Sbjct 182 KRTYWFDI-------YKNVEKISFVKCPVLVIHGTSDDVVNWSHGKQLFELC 226
> ath:AT5G14390 hypothetical protein
Length=369
Score = 49.7 bits (117), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 61/267 (22%), Positives = 94/267 (35%), Gaps = 73/267 (27%)
Query 59 KFPHIIGSPASYGIPYDDLF------------------LTTKDGVRIHGWFLKQTENEKA 100
KF SP+SY + YD+L L T+ G I +++
Sbjct 11 KFAFFPPSPSSYKLVYDELTGLLLMNPFPHRENVEILKLPTRRGTEIVAMYVRHPM--AT 68
Query 101 PTFILFHGNYGHVGLTLPRARWL-YDQGMNVLVIDYRGYGRSEGTPSEAGVYMDADAALD 159
T + HGN +G L +N++ DY GYG+S G PSE Y D +AA
Sbjct 69 STLLYSHGNAADLGQMYELFIELSIHLKVNLMGYDYSGYGQSTGKPSEHHTYADIEAAYK 128
Query 160 FVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGA 219
+ + +K + + G S+G
Sbjct 129 CL-------------------------------------EETYGAKQEDIILYGQSVGSG 151
Query 220 VAIDLAKRRGNELAGLVVENTFTSLREA--AEDTYAVFRLFRWLVKAIQRISMNNISKVG 277
+DLA R A ++ + LR + TY F +F+ NI K+
Sbjct 152 PTLDLAARLPQLRAAVLHSPILSGLRVMYPVKKTYW-FDIFK------------NIDKIP 198
Query 278 SLELPVLFLCGLRDENIKPRHSSRLYE 304
+ PVL + G DE + H +L+E
Sbjct 199 LVNCPVLVIHGTCDEVVDCSHGKQLWE 225
> ath:AT3G23540 hypothetical protein
Length=423
Score = 47.8 bits (112), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 61/121 (50%), Gaps = 20/121 (16%)
Query 218 GAVAIDLAKRRGNELAGLVVENTFTSLREAAE---DTYAVFRLFRWLVK--------AIQ 266
GAV + +AG+++++ F+ L + DTY FRL ++ VK AIQ
Sbjct 2 GAVTSLMYGVEDPSIAGMILDSPFSDLVDLMMELVDTYK-FRLPKFTVKFAIQFMRRAIQ 60
Query 267 R------ISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQKFLVEVPEGD 320
+ + +N I + +PVLF L D+ I+P HS R+YEA K +++ P GD
Sbjct 61 KKAKFDIMELNTIKVAKASFVPVLFGHALDDDFIRPHHSDRIYEAY-VGDKNIIKFP-GD 118
Query 321 H 321
H
Sbjct 119 H 119
> tgo:TGME49_061500 hypothetical protein
Length=501
Score = 44.7 bits (104), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 57/236 (24%), Positives = 97/236 (41%), Gaps = 33/236 (13%)
Query 90 WFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYD-QGMNVLVIDYRGYGRSEGTPSEA 148
+F++ E T + +HGN +G L +VL I++ GYG +
Sbjct 69 FFIEAPGGESQCTILYWHGNSCDLGQIYEEMDVLSKFLNAHVLAIEFPGYGLAPP----- 123
Query 149 GVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNF 208
++ D ++ SS + A + ++K Q G+ + S S+ NF
Sbjct 124 ---LNGPGPEDLAAAAIRAESSGEA-----APRRTTSGLAKNQMGELINKWSRSAF--NF 173
Query 209 ----------VFVMGHSMGGAVAIDLAKRRGNE---LAGLVVENTFTSLREAAEDTYAVF 255
V G S+G A LA E + G+V+ + ++ + ++ YA
Sbjct 174 LIWLGVAPASVICFGRSIGTGPASYLAAALAEENVHIGGVVLHAPYITVHKIVQE-YA-- 230
Query 256 RLFRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYEACGSSQK 311
L WL+ + N+ K+G+ P+L + GL DE I H RL+EA S +K
Sbjct 231 SLGTWLISN-HWSNAANLEKMGAASCPLLIVHGLDDEVIPTSHGRRLFEAYKSEKK 285
> ath:AT4G14290 hypothetical protein
Length=558
Score = 44.7 bits (104), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 32/116 (27%), Positives = 61/116 (52%), Gaps = 19/116 (16%)
Query 207 NFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAE---DTYAVFRLFRWLVK 263
+ + + G SMG ++ + +A +V+++ F+ L + DTY FRL ++ +K
Sbjct 135 SLIGLWGRSMGAVTSL-MYGAEDPSIAAMVLDSPFSDLVDLMMELVDTYK-FRLPKFTIK 192
Query 264 --------AIQRISMNNISKVGSLEL------PVLFLCGLRDENIKPRHSSRLYEA 305
A+Q+ + NI+ + ++++ PVLF + D+ I+P HS R+YEA
Sbjct 193 FAIQYMRRAVQKKANFNITDLNTIKVAKSCFVPVLFGHAVDDDFIQPHHSERIYEA 248
> dre:437017 zgc:100937
Length=166
Score = 43.5 bits (101), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 34/99 (34%), Positives = 47/99 (47%), Gaps = 3/99 (3%)
Query 76 DLFLT-TKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVI 133
+ F+T T G RI F++ + N + T + HGN +G L + NV
Sbjct 67 ECFMTRTSRGNRIACMFVRCSPNARY-TLLFSHGNAVDLGQMSSFYIGLGSRINCNVFSY 125
Query 134 DYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSS 172
DY GYG S G PSE +Y D DAA + + SS S+
Sbjct 126 DYSGYGASSGKPSEKNLYADVDAAWHALRTRSMISSPSA 164
> ath:AT2G24320 hypothetical protein
Length=286
Score = 42.0 bits (97), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 53/227 (23%), Positives = 83/227 (36%), Gaps = 51/227 (22%)
Query 79 LTTKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVIDYRG 137
LTTK G ++ F K + T + HGN +G + L +N++ DY G
Sbjct 41 LTTKSGNKVIATFWKHPFSRF--TLLYSHGNAADLGQMVDLFIELRAHLRVNIMSYDYSG 98
Query 138 YGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSS 197
YG S G P+E Y D +A + + I +++
Sbjct 99 YGASTGKPTELNTYYDIEAVYNCL--------------------RTEYGIMQEE------ 132
Query 198 SSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAVFRL 257
+ + G S+G + LA R L G+V+ + S Y V
Sbjct 133 -----------MILYGQSVGSGPTLHLASRV-KRLRGIVLHSAILS---GLRVLYPVKMT 177
Query 258 FRWLVKAIQRISMNNISKVGSLELPVLFLCGLRDENIKPRHSSRLYE 304
F W NI K+ + PVL + G +D+ + H RL+E
Sbjct 178 F-WFD------MYKNIDKIRHVTCPVLVIHGTKDDIVNMSHGKRLWE 217
> ath:AT4G31020 hypothetical protein
Length=294
Score = 41.2 bits (95), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 62/265 (23%), Positives = 90/265 (33%), Gaps = 69/265 (26%)
Query 59 KFPHIIGSPASYGIPYDD-----LF-------------LTTKDGVRIHGWFLKQTENEKA 100
KF PA+YG+ DD +F LTTK G ++ F +
Sbjct 11 KFAFFPPEPATYGVTKDDETGKLVFAGVSADKNVEVHQLTTKSGNKVVATFWRHPFARF- 69
Query 101 PTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVIDYRGYGRSEGTPSEAGVYMDADAALD 159
T + HGN +G + L +N++ DY GYG S G PSE Y D +A
Sbjct 70 -TLLYSHGNAADLGQMVELFIELRAHLRVNIMSYDYSGYGASTGKPSEFNTYYDIEAVY- 127
Query 160 FVLGKQQSSSSSSSNSNSNANNNNANAISKQQKGKSSSSSSSSSSKSNFVFVMGHSMGGA 219
S S K + + G S+G
Sbjct 128 ------------------------------------SCLRSDYGIKQEEIILYGQSVGSG 151
Query 220 VAIDLAKRRGNELAGLVVENTFTSLREAAEDTYAVFRLFRWLVKAIQRISMNNISKVGSL 279
+ +A R L G+V+ + S Y V ++ W NI K+ +
Sbjct 152 PTLHMASRL-KRLRGVVLHSAILS---GIRVLYPV-KMTLWF------DIFKNIDKIRHV 200
Query 280 ELPVLFLCGLRDENIKPRHSSRLYE 304
VL + G DE + H RL+E
Sbjct 201 NSQVLVIHGTNDEIVDLSHGKRLWE 225
> ath:AT2G39400 hydrolase, alpha/beta fold family protein
Length=311
Score = 40.4 bits (93), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 44/195 (22%), Positives = 79/195 (40%), Gaps = 34/195 (17%)
Query 72 IPYDDLFLTTKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTL-PRARWLYDQGMNV 130
+ Y++ F+ G+++ K + E L HG +T+ A L + G V
Sbjct 2 VMYEEDFVLNSRGMKLFTCVWKPVKQEPKALLFLCHGYAMESSITMNSAATRLANAGFAV 61
Query 131 LVIDYRGYGRSEGTPSEAGVYMDADAALDFVLGKQQSSSSSSSNSNSNANNNNANAISKQ 190
+DY G+G+SEG G + D +D V +N+ + I ++
Sbjct 62 YGMDYEGHGKSEGL---NGYISNFDDLVDDV-------------------SNHYSTICER 99
Query 191 QKGKSSSSSSSSSSKSNFVFVMGHSMGGAVAIDLAKRRGNELAGLVVENTFTSLREAAED 250
++ +K F++G SMGGAV + LA+++ + G V+ L + +
Sbjct 100 EE-----------NKGKMRFLLGESMGGAVVLLLARKKPDFWDGAVLVAPMCKLADEIKP 148
Query 251 TYAVFRLFRWLVKAI 265
V + L K I
Sbjct 149 HPVVISILIKLAKFI 163
Lambda K H
0.314 0.131 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 18188452224
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40