bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2872_orf1
Length=206
Score E
Sequences producing significant alignments: (Bits) Value
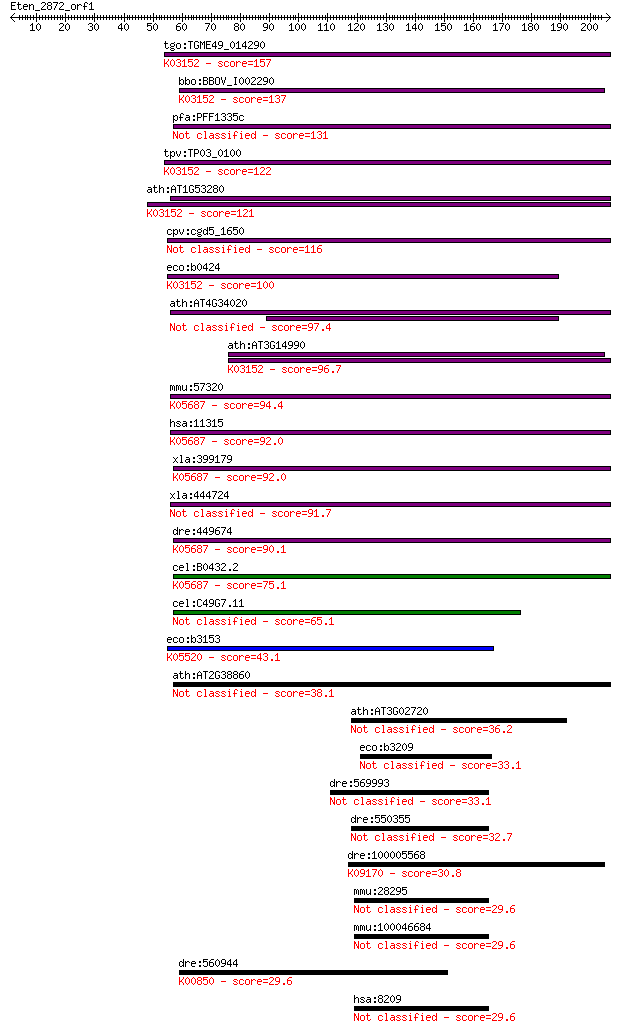
tgo:TGME49_014290 intracellular protease, putative ; K03152 4-... 157 3e-38
bbo:BBOV_I002290 19.m02374; 4-methyl-5(B-hydroxyethyl)-thiazol... 137 3e-32
pfa:PFF1335c 4-methyl-5(B-hydroxyethyl)-thiazol monophosphate ... 131 2e-30
tpv:TP03_0100 4-methyl-5(b-hydroxyethyl)-thiazole monophosphat... 122 1e-27
ath:AT1G53280 DJ-1 family protein; K03152 4-methyl-5(b-hydroxy... 121 1e-27
cpv:cgd5_1650 hypothetical protein 116 6e-26
eco:b0424 yajL, ECK0418, JW5057, thiJ; Oxidative-stress-resist... 100 4e-21
ath:AT4G34020 DJ-1 family protein 97.4 3e-20
ath:AT3G14990 4-methyl-5(b-hydroxyethyl)-thiazole monophosphat... 96.7 5e-20
mmu:57320 Park7, DJ-1, Dj1; Parkinson disease (autosomal reces... 94.4 2e-19
hsa:11315 PARK7, DJ-1, DJ1, FLJ27376, FLJ34360, FLJ92274; Park... 92.0 1e-18
xla:399179 park7, MGC82229, park7a; Parkinson disease (autosom... 92.0 1e-18
xla:444724 park7b; Parkinson disease (autosomal recessive, ear... 91.7 2e-18
dre:449674 park7, dj1, zgc:103725; parkinson disease (autosoma... 90.1 4e-18
cel:B0432.2 djr-1.1; DJ-1 (mammalian transcriptional regulator... 75.1 2e-13
cel:C49G7.11 djr-1.2; DJ-1 (mammalian transcriptional regulato... 65.1 2e-10
eco:b3153 yhbO, ECK3141, JW5529; stress-resistance protein; K0... 43.1 7e-04
ath:AT2G38860 YLS5; YLS5 38.1 0.023
ath:AT3G02720 DJ-1 family protein / protease-related 36.2 0.084
eco:b3209 elbB, ECK3198, elb2, JW3176, yhbL, yzzB; isoprenoid ... 33.1 0.72
dre:569993 MGC162944; zgc:162944 33.1 0.73
dre:550355 zgc:112056 32.7 0.93
dre:100005568 novel protein similar to vertebrate nuclear fact... 30.8 3.0
mmu:28295 D10Jhu81e, C21orf33, ES1; DNA segment, Chr 10, Johns... 29.6 7.2
mmu:100046684 ES1 protein homolog, mitochondrial-like 29.6 7.2
dre:560944 6-phosphofructokinase type C-like; K00850 6-phospho... 29.6 7.4
hsa:8209 C21orf33, ES1, GT335, HES1, KNPH, KNPI; chromosome 21... 29.6 7.5
> tgo:TGME49_014290 intracellular protease, putative ; K03152
4-methyl-5(b-hydroxyethyl)-thiazole monophosphate biosynthesis
Length=256
Score = 157 bits (396), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 78/155 (50%), Positives = 104/155 (67%), Gaps = 3/155 (1%)
Query 54 KMTKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKE 113
KM LV +A+ +E++E V+ +D LRRAG +V+VASV D+ V+ S G+ ++ D L+
Sbjct 71 KMAVKVLVPVAHDSEEIEAVSIIDTLRRAGAEVVVASVEDTEIVRMSRGVCVKADKLISA 130
Query 114 VSEQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLE 173
V E YD I IPGG+ GAE CRDS+ L A+L+ + K IAAICASPA+VLQ G L+
Sbjct 131 V-ENETYDCIAIPGGMPGAERCRDSAALTAMLKTHKAQGKLIAAICASPAVVLQTHGLLQ 189
Query 174 NVRAVAYPSFQQQLP--LKGEGRVCVDKHFITSVG 206
+AVAYP F Q P ++GEGRVCV +TSVG
Sbjct 190 GEKAVAYPCFMDQFPADMRGEGRVCVSNKIVTSVG 224
> bbo:BBOV_I002290 19.m02374; 4-methyl-5(B-hydroxyethyl)-thiazol
monophosphate biosynthesis enzyme; K03152 4-methyl-5(b-hydroxyethyl)-thiazole
monophosphate biosynthesis
Length=195
Score = 137 bits (345), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 70/147 (47%), Positives = 98/147 (66%), Gaps = 2/147 (1%)
Query 59 ALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVSEQN 118
AL+ +ANG+ED+EFV VDVLRRAG+ V VASV S V +HG KI D + +V+++
Sbjct 14 ALIAVANGSEDIEFVTVVDVLRRAGVTVTVASVHKSKDVVMAHGTKITADAAIDDVAKK- 72
Query 119 NYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLEN-VRA 177
+D+IV+PGGL G+ +C +S+ L +L +++ AAICA+PA+VL G L+ A
Sbjct 73 TFDLIVVPGGLPGSTHCAESTTLIKMLNQHKDGNRYYAAICAAPAVVLAAGGILDKQTAA 132
Query 178 VAYPSFQQQLPLKGEGRVCVDKHFITS 204
VAYP F+ LP G+GRVCV +TS
Sbjct 133 VAYPGFEDALPYVGKGRVCVSGKCVTS 159
> pfa:PFF1335c 4-methyl-5(B-hydroxyethyl)-thiazol monophosphate
biosynthesis enzyme
Length=189
Score = 131 bits (329), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 69/150 (46%), Positives = 97/150 (64%), Gaps = 1/150 (0%)
Query 57 KTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVSE 116
KTALV +A+G+EDVE++ VDVLRRAG+ V ASV S V + D + +V
Sbjct 5 KTALVAVASGSEDVEYITVVDVLRRAGVHVTTASVEKSEQVCLQSKNVVLADTTISKV-R 63
Query 117 QNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENVR 176
N YDV+VIPGG+KG+ + S +L+ Q ++++ AAICA+P VL + +++V
Sbjct 64 NNIYDVLVIPGGMKGSNTISECSEFIDMLKEQKANNRLYAAICAAPETVLDRHSLIDDVE 123
Query 177 AVAYPSFQQQLPLKGEGRVCVDKHFITSVG 206
AVAYPSF++ G+GRVCV K+ ITSVG
Sbjct 124 AVAYPSFERNFKHIGKGRVCVSKNCITSVG 153
> tpv:TP03_0100 4-methyl-5(b-hydroxyethyl)-thiazole monophosphate
biosynthesis protein; K03152 4-methyl-5(b-hydroxyethyl)-thiazole
monophosphate biosynthesis
Length=216
Score = 122 bits (305), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 75/154 (48%), Positives = 101/154 (65%), Gaps = 2/154 (1%)
Query 54 KMTKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKE 113
K T +ALV +G ED+E V VDVLRRAG+ V+VASV DSL + +HG K+ D +
Sbjct 28 KTTLSALVPAGDGTEDIELVTLVDVLRRAGVSVVVASVSDSLNLVLAHGTKLTADDKVTN 87
Query 114 VSEQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLE 173
++ Q +D+I +PGGL GA NC +S L +L+ Q SS + AAICASPALV G L+
Sbjct 88 LT-QKTFDLIAVPGGLVGATNCANSVGLVRMLKDQKSSGRLYAAICASPALVFGDCGLLD 146
Query 174 N-VRAVAYPSFQQQLPLKGEGRVCVDKHFITSVG 206
+ AVA+P F+ +LPL G+GRV V + +TS G
Sbjct 147 DKTSAVAFPGFESKLPLVGKGRVHVSHNCVTSQG 180
> ath:AT1G53280 DJ-1 family protein; K03152 4-methyl-5(b-hydroxyethyl)-thiazole
monophosphate biosynthesis
Length=438
Score = 121 bits (304), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 70/153 (45%), Positives = 100/153 (65%), Gaps = 3/153 (1%)
Query 56 TKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVS 115
T LV +A+G+E++E VA +DVL+RA V+VA++G+SL V S +K+ D+LL E +
Sbjct 257 TPQILVPIADGSEEMEAVAIIDVLKRAKANVVVAALGNSLEVVASRKVKLVADVLLDE-A 315
Query 116 EQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENV 175
E+N+YD+IV+PGGL GAE S L +L+ Q S+K AICASPALV + G L+
Sbjct 316 EKNSYDLIVLPGGLGGAEAFASSEKLVNMLKKQAESNKPYGAICASPALVFEPHGLLKGK 375
Query 176 RAVAYPSFQQQLPLKG--EGRVCVDKHFITSVG 206
+A A+P+ +L + E RV VD + ITS G
Sbjct 376 KATAFPAMCSKLTDQSHIEHRVLVDGNLITSRG 408
Score = 109 bits (272), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 60/162 (37%), Positives = 92/162 (56%), Gaps = 4/162 (2%)
Query 48 LSASPPKMTKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEG 107
+SA+ TK L+ +A+G E E V +DVLRR G V VASV + + V HG+K+
Sbjct 44 ISATMSSSTKKVLIPVAHGTEPFEAVVMIDVLRRGGADVTVASVENQVGVDACHGIKMVA 103
Query 108 DILLKEVSEQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQ 167
D LL ++++ + +D+I++PGGL G E ++ L +++ Q + + AAIC +PAL
Sbjct 104 DTLLSDITD-SVFDLIMLPGGLPGGETLKNCKPLEKMVKKQDTDGRLNAAICCAPALAFG 162
Query 168 QQGFLENVRAVAYPSFQQQLPLKG---EGRVCVDKHFITSVG 206
G LE +A YP F ++L E RV +D +TS G
Sbjct 163 TWGLLEGKKATCYPVFMEKLAACATAVESRVEIDGKIVTSRG 204
> cpv:cgd5_1650 hypothetical protein
Length=185
Score = 116 bits (290), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 65/155 (41%), Positives = 90/155 (58%), Gaps = 4/155 (2%)
Query 55 MTKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEV 114
M K LV LANG E++EFV VD+LRRA + V +A GD V +HG+ I GD LL EV
Sbjct 1 MPKKVLVALANGFEEIEFVTPVDILRRADLNVTIAVSGDCKRVMGAHGITIMGDKLLDEV 60
Query 115 SEQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLEN 174
+ +YD+++ PGG+ A +L +LR IA+ICASP +V ++ G L +
Sbjct 61 LSE-DYDLVMCPGGMDCAIKLGSDQNLLKILRETKKKGGIIASICASPVIVFEKNGLLSD 119
Query 175 V-RAVAYPSFQQQL--PLKGEGRVCVDKHFITSVG 206
V +AV+YPS +L P VCV + +TS G
Sbjct 120 VEKAVSYPSMMNELNRPDSSNAAVCVSSNVVTSQG 154
> eco:b0424 yajL, ECK0418, JW5057, thiJ; Oxidative-stress-resistance
chaperone; K03152 4-methyl-5(b-hydroxyethyl)-thiazole
monophosphate biosynthesis
Length=196
Score = 100 bits (248), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 56/136 (41%), Positives = 82/136 (60%), Gaps = 3/136 (2%)
Query 55 MTKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGD--SLFVKTSHGLKIEGDILLK 112
M+ +ALV LA G+E+ E V +D+L R G+KV ASV +L + S G+K+ D L
Sbjct 1 MSASALVCLAPGSEETEAVTTIDLLVRGGIKVTTASVASDGNLAITCSRGVKLLADAPLV 60
Query 113 EVSEQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFL 172
EV++ YDVIV+PGG+KGAE RDS+ L ++ H S + +AAICA+PA VL
Sbjct 61 EVAD-GEYDVIVLPGGIKGAECFRDSTLLVETVKQFHRSGRIVAAICAAPATVLVPHDIF 119
Query 173 ENVRAVAYPSFQQQLP 188
+P+ + ++P
Sbjct 120 PIGNMTGFPTLKDKIP 135
> ath:AT4G34020 DJ-1 family protein
Length=437
Score = 97.4 bits (241), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 63/156 (40%), Positives = 88/156 (56%), Gaps = 9/156 (5%)
Query 56 TKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVS 115
T L+ +ANG+E VE V+ DVLRRA + V V+SV SL + G KI D L+ E +
Sbjct 251 TPRVLIPVANGSEAVELVSIADVLRRAKVDVTVSSVERSLRITAFQGTKIITDKLIGEAA 310
Query 116 EQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENV 175
E ++YD+I++PGG G+E + S L LLR QH S + A +S + VL + G L+
Sbjct 311 E-SSYDLIILPGGHTGSERLQKSKILKKLLREQHESGRIYGATNSS-STVLHKHGLLKEK 368
Query 176 RAVAYPS-----FQQQLPLKGEGRVCVDKHFITSVG 206
R YPS QQ+ E V +D + ITS+G
Sbjct 369 RTTVYPSESDEPMNQQMIEGAE--VVIDGNVITSLG 402
Score = 61.6 bits (148), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 53/100 (53%), Gaps = 1/100 (1%)
Query 89 ASVGDSLFVKTSHGLKIEGDILLKEVSEQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQ 148
A+ L V+ S G ++ D+L+ + ++Q YD++ +PGG+ GA RD L +++ Q
Sbjct 81 ATTKKKLEVEGSSGTRLLADVLISKCADQV-YDLVALPGGMPGAVRLRDCEILEKIMKRQ 139
Query 149 HSSSKWIAAICASPALVLQQQGFLENVRAVAYPSFQQQLP 188
+ AI +PA+ L G L R +P+F +LP
Sbjct 140 AEDKRLYGAISMAPAITLLPWGLLTRKRTTGHPAFFGKLP 179
> ath:AT3G14990 4-methyl-5(b-hydroxyethyl)-thiazole monophosphate
biosynthesis protein, putative; K03152 4-methyl-5(b-hydroxyethyl)-thiazole
monophosphate biosynthesis
Length=369
Score = 96.7 bits (239), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 55/131 (41%), Positives = 80/131 (61%), Gaps = 3/131 (2%)
Query 76 VDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVSEQNNYDVIVIPGGLKGAENC 135
VD+LRRA V++A+VG+SL V+ S K+ ++LL EV+E++ +D+IV+PGGL GA+
Sbjct 208 VDILRRAKANVVIAAVGNSLEVEGSRKAKLVAEVLLDEVAEKS-FDLIVLPGGLNGAQRF 266
Query 136 RDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENVRAVAYPSFQQQLPLKG--EG 193
L +LR Q ++K ICASPA V + G L+ +A +P +L K E
Sbjct 267 ASCEKLVNMLRKQAEANKPYGGICASPAYVFEPNGLLKGKKATTHPVVSDKLSDKSHIEH 326
Query 194 RVCVDKHFITS 204
RV VD + ITS
Sbjct 327 RVVVDGNVITS 337
Score = 95.5 bits (236), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 52/135 (38%), Positives = 76/135 (56%), Gaps = 5/135 (3%)
Query 76 VDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVSEQNNYDVIVIPGGLKGAENC 135
+ VLRR G V VASV + V HG+K+ D LL ++++ + +D+IV+PGGL G E
Sbjct 2 ITVLRRGGADVTVASVETQVGVDACHGIKMVADTLLSDITD-SVFDLIVLPGGLPGGETL 60
Query 136 RDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENVRAVAYPSFQQQLPLKG---- 191
++ L +++ Q S + AAIC +PAL L G LE +A YP F ++L
Sbjct 61 KNCKSLENMVKKQDSDGRLNAAICCAPALALGTWGLLEGKKATGYPVFMEKLAATCATAV 120
Query 192 EGRVCVDKHFITSVG 206
E RV +D +TS G
Sbjct 121 ESRVQIDGRIVTSRG 135
> mmu:57320 Park7, DJ-1, Dj1; Parkinson disease (autosomal recessive,
early onset) 7; K05687 protein DJ-1
Length=189
Score = 94.4 bits (233), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 62/158 (39%), Positives = 89/158 (56%), Gaps = 10/158 (6%)
Query 56 TKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVS 115
+K ALV+LA GAE++E V VDV+RRAG+KV VA + V+ S + I D L++
Sbjct 3 SKRALVILAKGAEEMETVIPVDVMRRAGIKVTVAGLAGKDPVQCSRDVMICPDTSLEDAK 62
Query 116 EQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASP-ALVLQQQGFLEN 174
Q YDV+V+PGG GA+N +S + +L+ Q S IAAICA P AL+ + GF
Sbjct 63 TQGPYDVVVLPGGNLGAQNLSESPMVKEILKEQESRKGLIAAICAGPTALLAHEVGF--G 120
Query 175 VRAVAYPSFQQQLPLKG------EGRVCVDKHFITSVG 206
+ +P + ++ + G E RV D +TS G
Sbjct 121 CKVTTHPLAKDKM-MNGSHYSYSESRVEKDGLILTSRG 157
> hsa:11315 PARK7, DJ-1, DJ1, FLJ27376, FLJ34360, FLJ92274; Parkinson
disease (autosomal recessive, early onset) 7; K05687
protein DJ-1
Length=189
Score = 92.0 bits (227), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 60/157 (38%), Positives = 90/157 (57%), Gaps = 8/157 (5%)
Query 56 TKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVS 115
+K ALV+LA GAE++E V VDV+RRAG+KV VA + V+ S + I D L++
Sbjct 3 SKRALVILAKGAEEMETVIPVDVMRRAGIKVTVAGLAGKDPVQCSRDVVICPDASLEDAK 62
Query 116 EQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASP-ALVLQQQGFLEN 174
++ YDV+V+PGG GA+N +S+ + +L+ Q + IAAICA P AL+ + GF
Sbjct 63 KEGPYDVVVLPGGNLGAQNLSESAAVKEILKEQENRKGLIAAICAGPTALLAHEIGF--G 120
Query 175 VRAVAYPSFQQQLPLKG-----EGRVCVDKHFITSVG 206
+ +P + ++ G E RV D +TS G
Sbjct 121 SKVTTHPLAKDKMMNGGHYTYSENRVEKDGLILTSRG 157
> xla:399179 park7, MGC82229, park7a; Parkinson disease (autosomal
recessive, early onset) 7; K05687 protein DJ-1
Length=189
Score = 92.0 bits (227), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 61/156 (39%), Positives = 89/156 (57%), Gaps = 8/156 (5%)
Query 57 KTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVSE 116
K ALV+LA GAE++E V DV+RRAG+KV VA + V+ S + + D L+E
Sbjct 4 KRALVILAKGAEEMETVIPTDVMRRAGIKVTVAGLSGKDPVQCSRDVMLCPDTSLEEART 63
Query 117 QNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASP-ALVLQQQGFLENV 175
Q YDV+V+PGG GA+N +S + +L+ Q + IAAICA P AL + G + +
Sbjct 64 QGPYDVVVLPGGNLGAQNLSESPVVKEVLKEQEAKKGLIAAICAGPTALTVHGVGIGKTI 123
Query 176 RAVAYPSFQQQL--PLK---GEGRVCVDKHFITSVG 206
+P + ++ P + E RV D++FITS G
Sbjct 124 --TTHPLAKDKIVNPDQYKYSEERVVKDENFITSRG 157
> xla:444724 park7b; Parkinson disease (autosomal recessive, early
onset) 7 b
Length=189
Score = 91.7 bits (226), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 60/157 (38%), Positives = 90/157 (57%), Gaps = 8/157 (5%)
Query 56 TKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVS 115
+K ALV+LA GAE+ E V DV+RRAG+KV +A + V+ S + + D L+E
Sbjct 3 SKRALVILAKGAEETETVIPADVMRRAGIKVTIAGLNGKDPVQCSRDVMLCPDTSLEEAR 62
Query 116 EQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASP-ALVLQQQGFLEN 174
Q YDV+V+PGG GA+N +S + +L+ Q + IAAICA P AL + G ++
Sbjct 63 TQGPYDVVVLPGGNLGAQNLSESPVVKEVLKEQEAKKGLIAAICAGPTALTVHGVGIGKS 122
Query 175 VRAVAYPSFQQQL--PLK---GEGRVCVDKHFITSVG 206
+ +P + ++ P + E RV D++FITS G
Sbjct 123 I--TTHPLAKDKIVNPDQYKYSEERVVKDENFITSRG 157
> dre:449674 park7, dj1, zgc:103725; parkinson disease (autosomal
recessive, early onset) 7; K05687 protein DJ-1
Length=189
Score = 90.1 bits (222), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 59/156 (37%), Positives = 85/156 (54%), Gaps = 8/156 (5%)
Query 57 KTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVSE 116
K ALV+LA GAE++E V VDV+RRAG+ V VA + V+ S + I D L++ +
Sbjct 4 KRALVILAKGAEEMETVIPVDVMRRAGIAVTVAGLAGKEPVQCSREVMICPDSSLEDAHK 63
Query 117 QNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENVR 176
Q YDV+++PGGL GA+N +S + +L+ Q IAAICA P +L G
Sbjct 64 QGPYDVVLLPGGLLGAQNLSESPAVKEVLKDQEGRKGLIAAICAGPTALL-AHGIAYGST 122
Query 177 AVAYPSFQQQLPLKG------EGRVCVDKHFITSVG 206
+P + ++ + G E RV D + ITS G
Sbjct 123 VTTHPGAKDKM-MAGDHYKYSEARVQKDGNVITSRG 157
> cel:B0432.2 djr-1.1; DJ-1 (mammalian transcriptional regulator)
Related family member (djr-1.1); K05687 protein DJ-1
Length=187
Score = 75.1 bits (183), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 53/155 (34%), Positives = 86/155 (55%), Gaps = 8/155 (5%)
Query 57 KTALVVLA-NGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVS 115
K+AL++LA GAE++E + DVL R ++V+ A + + VK + G I D+ L++V
Sbjct 4 KSALIILAAEGAEEMEVIITGDVLARGEIRVVYAGLDGAEPVKCARGAHIVPDVKLEDV- 62
Query 116 EQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENV 175
E +D++++PGG G+ +S + +L++Q S I AICA+P +L E V
Sbjct 63 ETEKFDIVILPGGQPGSNTLAESLLVRDVLKSQVESGGLIGAICAAPIALLSHGVKAELV 122
Query 176 RAVAYPSFQQQLPLKG----EGRVCVDKHFITSVG 206
++PS +++L G E RV V ITS G
Sbjct 123 --TSHPSVKEKLEKGGYKYSEDRVVVSGKIITSRG 155
> cel:C49G7.11 djr-1.2; DJ-1 (mammalian transcriptional regulator)
Related family member (djr-1.2)
Length=186
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 43/120 (35%), Positives = 65/120 (54%), Gaps = 4/120 (3%)
Query 57 KTALVVLA-NGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVS 115
K+AL++L AE++E + DVL R G++VL A + VK + G +I D+ LK+V
Sbjct 5 KSALILLPPEDAEEIEVIVTGDVLVRGGLQVLYAG-SSTEPVKCAKGARIVPDVALKDV- 62
Query 116 EQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENV 175
+ +D+I+IPGG G + + LL+ Q S I AICA P ++L E V
Sbjct 63 KNKTFDIIIIPGG-PGCSKLAECPVIGELLKTQVKSGGLIGAICAGPTVLLAHGIVAERV 121
> eco:b3153 yhbO, ECK3141, JW5529; stress-resistance protein;
K05520 protease I [EC:3.2.-.-]
Length=172
Score = 43.1 bits (100), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 32/115 (27%), Positives = 54/115 (46%), Gaps = 7/115 (6%)
Query 55 MTKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILL-KE 113
M+K V++ + ED EF + D R+AG +V+ + KT G K E + + K
Sbjct 1 MSKKIAVLITDEFEDSEFTSPADEFRKAGHEVITI---EKQAGKTVKGKKGEASVTIDKS 57
Query 114 VSEQN--NYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVL 166
+ E +D +++PGG + R + R +S K + AIC P L++
Sbjct 58 IDEVTPAEFDALLLPGG-HSPDYLRGDNRFVTFTRDFVNSGKPVFAICHGPQLLI 111
> ath:AT2G38860 YLS5; YLS5
Length=279
Score = 38.1 bits (87), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 47/184 (25%), Positives = 79/184 (42%), Gaps = 41/184 (22%)
Query 57 KTALVVLANGAEDVEFVAAVDVLRRAGMKVLVAS----VGDSLFVKTSH---GLKIEGDI 109
K+AL++ + E E + + VL+ G+ V S GD V ++H GL++ ++
Sbjct 7 KSALLLCGDYMEAYETIVPLYVLQSFGVSVHCVSPNRNAGDRC-VMSAHDFLGLELYTEL 65
Query 110 LLKEVS--------EQNNYDVIVIPGG-----LKGAENCRDSSHLAALLRAQHSSSKWIA 156
++ +++ NYDVI+IPGG L E C D L+ S K I
Sbjct 66 VVDQLTLNANFDDVTPENYDVIIIPGGRFTELLSADEKCVD------LVARFAESKKLIF 119
Query 157 AICASPALVLQQQGFLENVRAVAYPSFQQQLPLKG-------------EGRVCV-DKHFI 202
C S +++ V+ A+ S + + L G E CV D +F+
Sbjct 120 TSCHSQVMLMAAGILAGGVKCTAFESIKPLIELSGGEWWQQPGIQSMFEITDCVKDGNFM 179
Query 203 TSVG 206
++VG
Sbjct 180 STVG 183
> ath:AT3G02720 DJ-1 family protein / protease-related
Length=388
Score = 36.2 bits (82), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 40/74 (54%), Gaps = 2/74 (2%)
Query 118 NNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENVRA 177
++YD +VIPGG + E + H+ +++ +S K +A+IC +L G L+ +
Sbjct 273 SSYDALVIPGG-RAPEYLALNEHVLNIVKEFMNSEKPVASICHGQQ-ILAAAGVLKGRKC 330
Query 178 VAYPSFQQQLPLKG 191
AYP+ + + L G
Sbjct 331 TAYPAVKLNVVLGG 344
> eco:b3209 elbB, ECK3198, elb2, JW3176, yhbL, yzzB; isoprenoid
biosynthesis protein with amidotransferase-like domain
Length=217
Score = 33.1 bits (74), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 27/55 (49%), Gaps = 10/55 (18%)
Query 121 DVIVIPGGLKGAEN----------CRDSSHLAALLRAQHSSSKWIAAICASPALV 165
D +++PGG A+N C L AL +A H + K + +C +PA++
Sbjct 87 DALIVPGGFGAAKNLSNFASLGSECTVDRELKALAQAMHQAGKPLGFMCIAPAML 141
> dre:569993 MGC162944; zgc:162944
Length=224
Score = 33.1 bits (74), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 33/66 (50%), Gaps = 12/66 (18%)
Query 111 LKEVSEQN--NYDVIVIPGGLKGAEN----------CRDSSHLAALLRAQHSSSKWIAAI 158
++++S+ N + D I+ PGG A+N C + + A+L+A H+ K I
Sbjct 76 IEDLSQLNVKDLDAIIFPGGFGAAKNLCSWAVDGKDCSVNDQVKAVLQAFHAEKKPIGLC 135
Query 159 CASPAL 164
C SP L
Sbjct 136 CISPVL 141
> dre:550355 zgc:112056
Length=255
Score = 32.7 bits (73), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 26/57 (45%), Gaps = 10/57 (17%)
Query 118 NNYDVIVIPGGLKGAEN----------CRDSSHLAALLRAQHSSSKWIAAICASPAL 164
+N+D I+ PGG A+N C + + +L+ H + K I C SP L
Sbjct 113 SNHDAIIFPGGFGAAKNLSTFAIDGMDCEVNKEVERVLKDFHKAGKPIGLCCISPVL 169
> dre:100005568 novel protein similar to vertebrate nuclear factor
I/C (CCAAT-binding transcription factor) (NFIC); K09170
nuclear factor I/C
Length=553
Score = 30.8 bits (68), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 39/89 (43%), Gaps = 1/89 (1%)
Query 117 QNNYDVIVIPGGLKGAENCRDSSHLAAL-LRAQHSSSKWIAAICASPALVLQQQGFLENV 175
Q++ VI + G+ + + S H + Q +S+ + P + Q + V
Sbjct 369 QHHRPVIAVHSGISRSPHPSSSLHFPGTPILPQTASTYFPHTAIRYPPHLSSQDPLKDLV 428
Query 176 RAVAYPSFQQQLPLKGEGRVCVDKHFITS 204
PS QQ PL G G+V V H+I+S
Sbjct 429 SLACDPSNQQPSPLNGSGQVKVSSHYISS 457
> mmu:28295 D10Jhu81e, C21orf33, ES1; DNA segment, Chr 10, Johns
Hopkins University 81 expressed
Length=266
Score = 29.6 bits (65), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 15/56 (26%), Positives = 25/56 (44%), Gaps = 10/56 (17%)
Query 119 NYDVIVIPGGLKGAEN----------CRDSSHLAALLRAQHSSSKWIAAICASPAL 164
N+D + PGG A+N C+ + + +L+ H + K I C +P L
Sbjct 125 NHDAAIFPGGFGAAKNLSTFAVDGKDCKVNKEVERVLKEFHGAKKPIGLCCIAPVL 180
> mmu:100046684 ES1 protein homolog, mitochondrial-like
Length=266
Score = 29.6 bits (65), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 15/56 (26%), Positives = 25/56 (44%), Gaps = 10/56 (17%)
Query 119 NYDVIVIPGGLKGAEN----------CRDSSHLAALLRAQHSSSKWIAAICASPAL 164
N+D + PGG A+N C+ + + +L+ H + K I C +P L
Sbjct 125 NHDAAIFPGGFGAAKNLSTFAVDGKDCKVNKEVERVLKEFHGAKKPIGLCCIAPVL 180
> dre:560944 6-phosphofructokinase type C-like; K00850 6-phosphofructokinase
[EC:2.7.1.11]
Length=785
Score = 29.6 bits (65), Expect = 7.4, Method: Composition-based stats.
Identities = 23/92 (25%), Positives = 42/92 (45%), Gaps = 4/92 (4%)
Query 59 ALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVSEQN 118
A+ VL +G + AAV + R G+ V FV + ++G +KE + +N
Sbjct 19 AIAVLTSGGDAQGMNAAVRAVVRMGIYV----GAKVYFVHEGYQGMVDGGDNIKEATWEN 74
Query 119 NYDVIVIPGGLKGAENCRDSSHLAALLRAQHS 150
++ + G + G+ C+D L+A H+
Sbjct 75 VSSMLQVGGTVIGSARCKDFRTHEGRLKAAHN 106
> hsa:8209 C21orf33, ES1, GT335, HES1, KNPH, KNPI; chromosome
21 open reading frame 33
Length=268
Score = 29.6 bits (65), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 15/56 (26%), Positives = 25/56 (44%), Gaps = 10/56 (17%)
Query 119 NYDVIVIPGGLKGAEN----------CRDSSHLAALLRAQHSSSKWIAAICASPAL 164
N+D + PGG A+N C+ + + +L+ H + K I C +P L
Sbjct 127 NHDAAIFPGGFGAAKNLSTFAVDGKDCKVNKEVERVLKEFHQAGKPIGLCCIAPVL 182
Lambda K H
0.316 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6382561284
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40