bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2875_orf2
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
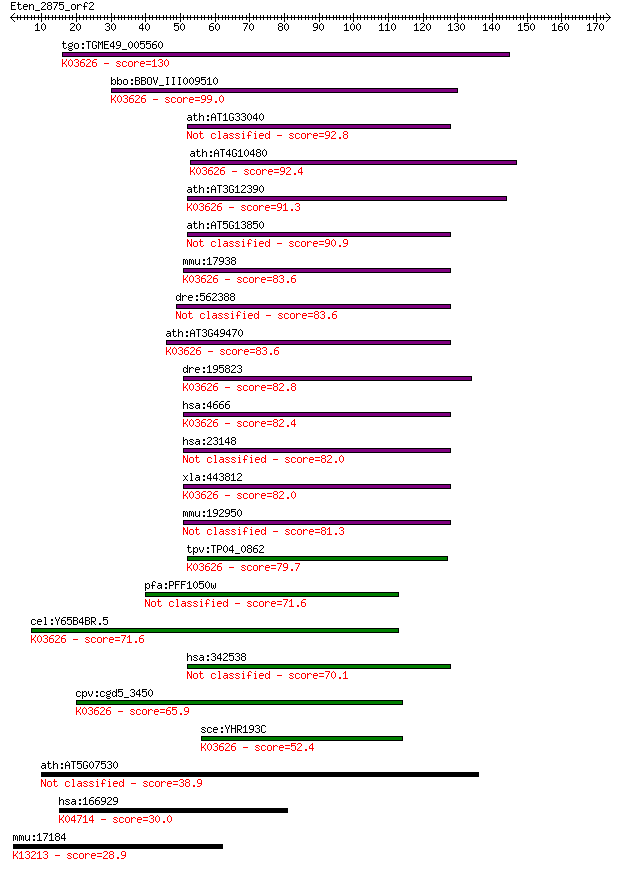
tgo:TGME49_005560 nascent polypeptide-associated complex alpha... 130 3e-30
bbo:BBOV_III009510 17.m07825; NAC domain containing protein; K... 99.0 7e-21
ath:AT1G33040 NACA5; NACA5 (NASCENT POLYPEPTIDE-ASSOCIATED COM... 92.8 4e-19
ath:AT4G10480 nascent polypeptide associated complex alpha cha... 92.4 7e-19
ath:AT3G12390 nascent polypeptide associated complex alpha cha... 91.3 1e-18
ath:AT5G13850 NACA3; NACA3 (NASCENT POLYPEPTIDE-ASSOCIATED COM... 90.9 2e-18
mmu:17938 Naca, AL022831, AL024382, Gm1878, mKIAA0363, skNAC; ... 83.6 3e-16
dre:562388 tbrg4; transforming growth factor beta regulator 4 83.6
ath:AT3G49470 NACA2; NACA2 (NASCENT POLYPEPTIDE-ASSOCIATED COM... 83.6 3e-16
dre:195823 naca, ik:tdsubc_2b5, nkn, tdsubc_2b5, xx:tdsubc_2b5... 82.8 5e-16
hsa:4666 NACA, MGC117224, NACA1; nascent polypeptide-associate... 82.4 8e-16
hsa:23148 NACAD, KIAA0363; NAC alpha domain containing 82.0 8e-16
xla:443812 naca.2, naca, naca1; nascent polypeptide-associated... 82.0 1e-15
mmu:192950 Nacad, D230024G13Rik, KIAA0363, mKIAA0363; NAC alph... 81.3 2e-15
tpv:TP04_0862 nascent polypeptide associated complex subunit a... 79.7 4e-15
pfa:PFF1050w nascent polypeptide associated complex alpha chai... 71.6 1e-12
cel:Y65B4BR.5 hypothetical protein; K03626 nascent polypeptide... 71.6 1e-12
hsa:342538 NACA2, ANAC, MGC71999, NACAL; nascent polypeptide-a... 70.1 4e-12
cpv:cgd5_3450 nascent polypeptide associated complex alpha cha... 65.9 6e-11
sce:YHR193C EGD2; Alpha subunit of the heteromeric nascent pol... 52.4 8e-07
ath:AT5G07530 GRP17; GRP17 (GLYCINE RICH PROTEIN 17); lipid bi... 38.9 0.008
hsa:166929 SGMS2, MGC26963, SMS2; sphingomyelin synthase 2 (EC... 30.0 3.8
mmu:17184 Matr3, 1110061A14Rik, 2810017I02Rik, AI841759, AW555... 28.9 7.9
> tgo:TGME49_005560 nascent polypeptide-associated complex alpha
chain, putative (EC:3.4.21.68 3.6.3.8); K03626 nascent polypeptide-associated
complex subunit alpha
Length=6052
Score = 130 bits (326), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 74/132 (56%), Positives = 95/132 (71%), Gaps = 4/132 (3%)
Query 16 EEPEHISSSSSDSGSEDEQKGEGAQQEQKEGEGSPS-RSRQSRNERKARKAILRLGMKPM 74
EE +H+S+SS DS S DE+ + + Q+E G+P RS+QSRNE+KARKA+LR GMK M
Sbjct 5868 EERKHLSASSVDSES-DEECPKLEEVNQEEDAGTPEGRSKQSRNEKKARKAVLRHGMKHM 5926
Query 75 PDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGHA--EGVATQAHSEAAQRFTQGAV 132
P IV+V+IRK+K FV+ +PDVYKS ++D YVIFG A E + +QAHS AAQRFTQ A
Sbjct 5927 PGIVRVMIRKSKHILFVVPKPDVYKSLASDTYVIFGEAKIEDLNSQAHSAAAQRFTQAAN 5986
Query 133 SFGGGEGAAAAA 144
+ GAA AA
Sbjct 5987 AQFPANGAATAA 5998
> bbo:BBOV_III009510 17.m07825; NAC domain containing protein;
K03626 nascent polypeptide-associated complex subunit alpha
Length=207
Score = 99.0 bits (245), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 50/104 (48%), Positives = 71/104 (68%), Gaps = 4/104 (3%)
Query 30 SEDEQKGEGAQ--QEQKEGEGSPSRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQ 87
SEDE EG +E K +GS S+ RQ++NERK+RK + +LG+KP+ I KV I+K+KQ
Sbjct 17 SEDEVSSEGDSDIEESKGADGSSSKGRQNKNERKSRKLLGKLGLKPVEGITKVCIKKSKQ 76
Query 88 SWFVIAEPDVYKSTSTDAYVIFGHA--EGVATQAHSEAAQRFTQ 129
+FV+ +PDVYK ++D Y+IFG A E + + EAAQR +Q
Sbjct 77 VFFVVNKPDVYKLPNSDTYIIFGEAKVEDMGQNSALEAAQRLSQ 120
> ath:AT1G33040 NACA5; NACA5 (NASCENT POLYPEPTIDE-ASSOCIATED COMPLEX
SUBUNIT ALPHA-LIKE PROTEIN 5)
Length=209
Score = 92.8 bits (229), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 40/78 (51%), Positives = 62/78 (79%), Gaps = 2/78 (2%)
Query 52 RSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGH 111
++QSR+E+K+RKA+L+LGMKP+ D+ +V I++AK FVI++PDVYKS + + YVIFG
Sbjct 58 NAKQSRSEKKSRKAVLKLGMKPVSDVSRVTIKRAKNVLFVISKPDVYKSPNAETYVIFGE 117
Query 112 A--EGVATQAHSEAAQRF 127
A + +++Q ++AAQRF
Sbjct 118 AKVDDLSSQLQTQAAQRF 135
> ath:AT4G10480 nascent polypeptide associated complex alpha chain
protein, putative / alpha-NAC, putative; K03626 nascent
polypeptide-associated complex subunit alpha
Length=211
Score = 92.4 bits (228), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 44/98 (44%), Positives = 71/98 (72%), Gaps = 4/98 (4%)
Query 53 SRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGHA 112
S+QSR+E+K+RKA+L+LGMKP+ D+ +V I+++K FVI++PDV+KS +++ YVIFG A
Sbjct 61 SKQSRSEKKSRKAMLKLGMKPVTDVSRVTIKRSKNVLFVISKPDVFKSPNSETYVIFGEA 120
Query 113 --EGVATQAHSEAAQRFTQGAVS--FGGGEGAAAAAAA 146
+ +++Q ++AAQRF V+ +G+ AA A
Sbjct 121 KIDDMSSQLQAQAAQRFKMPDVASMIPNTDGSEAATVA 158
> ath:AT3G12390 nascent polypeptide associated complex alpha chain
protein, putative / alpha-NAC, putative; K03626 nascent
polypeptide-associated complex subunit alpha
Length=203
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 44/96 (45%), Positives = 71/96 (73%), Gaps = 4/96 (4%)
Query 52 RSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGH 111
+S+QSR+E+K+RKA+L+LGMKP+ + +V ++K+K FVI++PDV+KS ++D YVIFG
Sbjct 56 KSKQSRSEKKSRKAMLKLGMKPITGVSRVTVKKSKNILFVISKPDVFKSPASDTYVIFGE 115
Query 112 A--EGVATQAHSEAAQRFTQGAVS--FGGGEGAAAA 143
A E +++Q S+AA++F +S GE ++AA
Sbjct 116 AKIEDLSSQIQSQAAEQFKAPDLSNVISKGESSSAA 151
> ath:AT5G13850 NACA3; NACA3 (NASCENT POLYPEPTIDE-ASSOCIATED COMPLEX
SUBUNIT ALPHA-LIKE PROTEIN 3)
Length=204
Score = 90.9 bits (224), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 40/78 (51%), Positives = 63/78 (80%), Gaps = 2/78 (2%)
Query 52 RSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGH 111
RS+QSR+E+K+RKA+L+LGMKP+ + +V ++K+K FVI++PDV+KS ++D YVIFG
Sbjct 52 RSKQSRSEKKSRKAMLKLGMKPITGVSRVTVKKSKNILFVISKPDVFKSPASDTYVIFGE 111
Query 112 A--EGVATQAHSEAAQRF 127
A E +++Q S+AA++F
Sbjct 112 AKIEDLSSQLQSQAAEQF 129
> mmu:17938 Naca, AL022831, AL024382, Gm1878, mKIAA0363, skNAC;
nascent polypeptide-associated complex alpha polypeptide (EC:5.3.99.3);
K03626 nascent polypeptide-associated complex
subunit alpha
Length=2187
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 37/79 (46%), Positives = 59/79 (74%), Gaps = 2/79 (2%)
Query 51 SRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFG 110
S+++QSR+E+KARKA+ +LG++ + + +V IRK+K FVI +PDVYKS ++D Y++FG
Sbjct 2037 SKAKQSRSEKKARKAMSKLGLRQVTGVTRVTIRKSKNILFVITKPDVYKSPASDTYIVFG 2096
Query 111 HA--EGVATQAHSEAAQRF 127
A E ++ QA AA++F
Sbjct 2097 EAKIEDLSQQAQLAAAEKF 2115
> dre:562388 tbrg4; transforming growth factor beta regulator
4
Length=2317
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 38/81 (46%), Positives = 59/81 (72%), Gaps = 2/81 (2%)
Query 49 SPSRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVI 108
S SR++QSR+E+KARKA+ +LG+K + + ++ IRK+K FVI PDV+KS ++D Y++
Sbjct 2164 SVSRAKQSRSEKKARKAMSKLGLKQIHGVTRITIRKSKNILFVITRPDVFKSPASDIYIV 2223
Query 109 FGHA--EGVATQAHSEAAQRF 127
FG A E ++ Q H AA++F
Sbjct 2224 FGEAKIEDLSQQVHKAAAEKF 2244
> ath:AT3G49470 NACA2; NACA2 (NASCENT POLYPEPTIDE-ASSOCIATED COMPLEX
SUBUNIT ALPHA-LIKE PROTEIN 2); K03626 nascent polypeptide-associated
complex subunit alpha
Length=217
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 38/84 (45%), Positives = 61/84 (72%), Gaps = 2/84 (2%)
Query 46 GEGSPSRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDA 105
+G S+QSR+E+K+RKA+L+LGMKP+ + +V I++ K F I++PDV+KS ++
Sbjct 60 AQGVSGSSKQSRSEKKSRKAMLKLGMKPVTGVSRVTIKRTKNVLFFISKPDVFKSPHSET 119
Query 106 YVIFGHA--EGVATQAHSEAAQRF 127
YVIFG A E +++Q ++AAQ+F
Sbjct 120 YVIFGEAKIEDLSSQLQTQAAQQF 143
> dre:195823 naca, ik:tdsubc_2b5, nkn, tdsubc_2b5, xx:tdsubc_2b5;
nascent polypeptide-associated complex alpha polypeptide
(EC:5.3.99.3); K03626 nascent polypeptide-associated complex
subunit alpha
Length=1030
Score = 82.8 bits (203), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 42/87 (48%), Positives = 64/87 (73%), Gaps = 4/87 (4%)
Query 51 SRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFG 110
S+++QSR+E+KARKA+ +LG++ + + +V IRK+K FVI +PDVYKS ++D Y++FG
Sbjct 880 SKAKQSRSEKKARKAMSKLGLRQVAGVTRVTIRKSKNILFVITKPDVYKSPASDTYIVFG 939
Query 111 HA--EGVATQAHSEAAQRF-TQG-AVS 133
A E ++ QA AA++F QG AVS
Sbjct 940 EAKIEDLSQQAQLAAAEKFKVQGEAVS 966
> hsa:4666 NACA, MGC117224, NACA1; nascent polypeptide-associated
complex alpha subunit (EC:5.3.99.3); K03626 nascent polypeptide-associated
complex subunit alpha
Length=215
Score = 82.4 bits (202), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 37/79 (46%), Positives = 59/79 (74%), Gaps = 2/79 (2%)
Query 51 SRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFG 110
S+++QSR+E+KARKA+ +LG++ + + +V IRK+K FVI +PDVYKS ++D Y++FG
Sbjct 65 SKAKQSRSEKKARKAMSKLGLRQVTGVTRVTIRKSKNILFVITKPDVYKSPASDTYIVFG 124
Query 111 HA--EGVATQAHSEAAQRF 127
A E ++ QA AA++F
Sbjct 125 EAKIEDLSQQAQLAAAEKF 143
> hsa:23148 NACAD, KIAA0363; NAC alpha domain containing
Length=1562
Score = 82.0 bits (201), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 60/79 (75%), Gaps = 2/79 (2%)
Query 51 SRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFG 110
++++QSR+E+KARKA+ +LG++ + + ++ I+K+K FVIA+PDV+KS ++D YV+FG
Sbjct 1406 AKAKQSRSEKKARKAMSKLGLRQIQGVTRITIQKSKNILFVIAKPDVFKSPASDTYVVFG 1465
Query 111 HA--EGVATQAHSEAAQRF 127
A E ++ Q H AA++F
Sbjct 1466 EAKIEDLSQQVHKAAAEKF 1484
> xla:443812 naca.2, naca, naca1; nascent polypeptide-associated
complex alpha subunit (EC:5.3.99.3); K03626 nascent polypeptide-associated
complex subunit alpha
Length=213
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 37/79 (46%), Positives = 59/79 (74%), Gaps = 2/79 (2%)
Query 51 SRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFG 110
S+++QSR+E+KARKA+ +LG++ + + +V IRK+K FVI +PDVYKS ++D Y++FG
Sbjct 63 SKAKQSRSEKKARKAMSKLGLRQVTGVTRVTIRKSKNILFVITKPDVYKSPASDTYIVFG 122
Query 111 HA--EGVATQAHSEAAQRF 127
A E ++ QA AA++F
Sbjct 123 EAKIEDLSQQAQLAAAEKF 141
> mmu:192950 Nacad, D230024G13Rik, KIAA0363, mKIAA0363; NAC alpha
domain containing
Length=1504
Score = 81.3 bits (199), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 60/79 (75%), Gaps = 2/79 (2%)
Query 51 SRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFG 110
++++QSR+E+KARKA+ +LG++ + + ++ I+K+K FVIA+PDV+KS ++D YV+FG
Sbjct 1349 AKAKQSRSEKKARKAMSKLGLRQIQGVTRITIQKSKNILFVIAKPDVFKSPASDTYVVFG 1408
Query 111 HA--EGVATQAHSEAAQRF 127
A E ++ Q H AA++F
Sbjct 1409 EAKIEDLSQQVHKAAAEKF 1427
> tpv:TP04_0862 nascent polypeptide associated complex subunit
alpha; K03626 nascent polypeptide-associated complex subunit
alpha
Length=200
Score = 79.7 bits (195), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 39/77 (50%), Positives = 55/77 (71%), Gaps = 2/77 (2%)
Query 52 RSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGH 111
+++ +NERKARKA+ +LG+K + K+ IRK+KQ +FVI +PDV+K ++D YVIFG
Sbjct 50 KTKVGKNERKARKAMSKLGLKQYEGVSKIYIRKSKQIFFVINKPDVFKLPNSDIYVIFGE 109
Query 112 A--EGVATQAHSEAAQR 126
A E V + SEAAQR
Sbjct 110 AKLEDVGANSQSEAAQR 126
> pfa:PFF1050w nascent polypeptide associated complex alpha chain,
putative
Length=184
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 51/73 (69%), Gaps = 0/73 (0%)
Query 40 QQEQKEGEGSPSRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYK 99
E K P+R++ S+ ER+ARK +++LG+K +P++ KV+I+K+++ F ++ +VYK
Sbjct 21 NDENKNILNPPNRTKMSKGERRARKMLVKLGLKAIPNVHKVIIKKSQKMVFAVSNVEVYK 80
Query 100 STSTDAYVIFGHA 112
TD+YVIFG A
Sbjct 81 VEGTDSYVIFGDA 93
> cel:Y65B4BR.5 hypothetical protein; K03626 nascent polypeptide-associated
complex subunit alpha
Length=197
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 42/117 (35%), Positives = 64/117 (54%), Gaps = 19/117 (16%)
Query 7 AAEMANKKAEEPEHISSSSSDSGSEDEQKGEGAQQEQKE-----------GEGSPSRSRQ 55
+ E K+ +EP+ S SD+ E +QE E G+ +++Q
Sbjct 4 STETRQKEVKEPQVDVSDDSDN--------EAVEQELTEEQRRVAEAAGLGDHIDKQAKQ 55
Query 56 SRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGHA 112
SR+E+KARK +LG+K + + +V IRK+K FVI +PDV+KS +D Y+IFG A
Sbjct 56 SRSEKKARKLFSKLGLKQVTGVSRVCIRKSKNILFVINKPDVFKSPGSDTYIIFGEA 112
> hsa:342538 NACA2, ANAC, MGC71999, NACAL; nascent polypeptide-associated
complex alpha subunit 2
Length=215
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 33/78 (42%), Positives = 56/78 (71%), Gaps = 2/78 (2%)
Query 52 RSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGH 111
+++QSR+E++ARKA+ +LG+ + + +V I K+K FVI + DVYKS ++DAY++FG
Sbjct 66 KAKQSRSEKRARKAMSKLGLLQVTGVTRVTIWKSKNILFVITKLDVYKSPASDAYIVFGE 125
Query 112 A--EGVATQAHSEAAQRF 127
A + ++ QA AA++F
Sbjct 126 AKIQDLSQQAQLAAAEKF 143
> cpv:cgd5_3450 nascent polypeptide associated complex alpha chain
with an NAC domain ; K03626 nascent polypeptide-associated
complex subunit alpha
Length=196
Score = 65.9 bits (159), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 37/94 (39%), Positives = 52/94 (55%), Gaps = 8/94 (8%)
Query 20 HISSSSSDSGSEDEQKGEGAQQEQKEGEGSPSRSRQSRNERKARKAILRLGMKPMPDIVK 79
H SS SEDE G + EG R R ++ E+K RK + +LG+KP+PD K
Sbjct 13 HDDQISSGESSEDEDLNSG----KTEG----VRPRHNKGEKKNRKLVQKLGLKPLPDFEK 64
Query 80 VVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGHAE 113
VV+R A+ F I P+VY T +Y++FG A+
Sbjct 65 VVVRGARGLSFQIVNPEVYCVPGTKSYIVFGDAK 98
> sce:YHR193C EGD2; Alpha subunit of the heteromeric nascent polypeptide-associated
complex (NAC) involved in protein sorting
and translocation, associated with cytoplasmic ribosomes;
K03626 nascent polypeptide-associated complex subunit alpha
Length=174
Score = 52.4 bits (124), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 40/58 (68%), Gaps = 1/58 (1%)
Query 56 SRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGHAE 113
++NE+KAR+ I +LG+K +P I++V RK + I +P+V++S + YV+FG A+
Sbjct 14 NKNEKKARELIGKLGLKQIPGIIRVTFRKKDNQIYAIEKPEVFRSAGGN-YVVFGEAK 70
> ath:AT5G07530 GRP17; GRP17 (GLYCINE RICH PROTEIN 17); lipid
binding
Length=512
Score = 38.9 bits (89), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 36/132 (27%), Positives = 56/132 (42%), Gaps = 10/132 (7%)
Query 10 MANKKAEEPEHISSSSSDSGSEDEQKGEGAQQEQKEGEGSPSRSRQSRNERKARKAILRL 69
M+ + +++P SS+ S S E + Q+EG PS S R+ RK +
Sbjct 1 MSEELSQKP---SSAQSLSLREGRNRFPFLSLSQREGRFFPSLSLSERDGRKFSFLSMFS 57
Query 70 GMKPMPDIVKVVIRKAKQSWFV------IAEPDVYKSTSTDAYVIFGHAEGVATQAHSEA 123
+ P+ +++K++I FV +A ST ++IF AT A
Sbjct 58 FLMPLLEVIKIIIASVASVIFVGFACVTLAGSAAALVVSTPVFIIFSPVLVPATIATVVL 117
Query 124 AQRFTQGAVSFG 135
A FT G SFG
Sbjct 118 ATGFTAGG-SFG 128
> hsa:166929 SGMS2, MGC26963, SMS2; sphingomyelin synthase 2 (EC:2.7.8.27);
K04714 shingomyelin synthase [EC:2.7.8.27]
Length=365
Score = 30.0 bits (66), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 30/66 (45%), Gaps = 10/66 (15%)
Query 15 AEEPEHISSSSSDSGSEDEQKGEGAQQEQKEGEGSPSRSRQSRNERKARKAILRLGMKPM 74
A+ EH+ + SD + + E ++E K G G P K+ + LR G K
Sbjct 7 AKLEEHLENQPSDPTNTYARPAEPVEEENKNGNGKP----------KSLSSGLRKGTKKY 56
Query 75 PDIVKV 80
PD +++
Sbjct 57 PDYIQI 62
> mmu:17184 Matr3, 1110061A14Rik, 2810017I02Rik, AI841759, AW555618,
D030046F20Rik, mKIAA0723; matrin 3; K13213 matrin 3
Length=846
Score = 28.9 bits (63), Expect = 7.9, Method: Composition-based stats.
Identities = 17/64 (26%), Positives = 30/64 (46%), Gaps = 4/64 (6%)
Query 2 GFFVLAAEMANKKAEEPEHISSSSSDSGSEDEQK----GEGAQQEQKEGEGSPSRSRQSR 57
G +L + + K++ P+ S S D QK EG +QE+K GE ++ +
Sbjct 583 GIDLLKKDKSRKRSYSPDGKESPSDKKSKTDAQKTESPAEGKEQEEKSGEDGEKDTKDDQ 642
Query 58 NERK 61
E++
Sbjct 643 TEQE 646
Lambda K H
0.310 0.123 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4406352944
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40