bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2892_orf1
Length=193
Score E
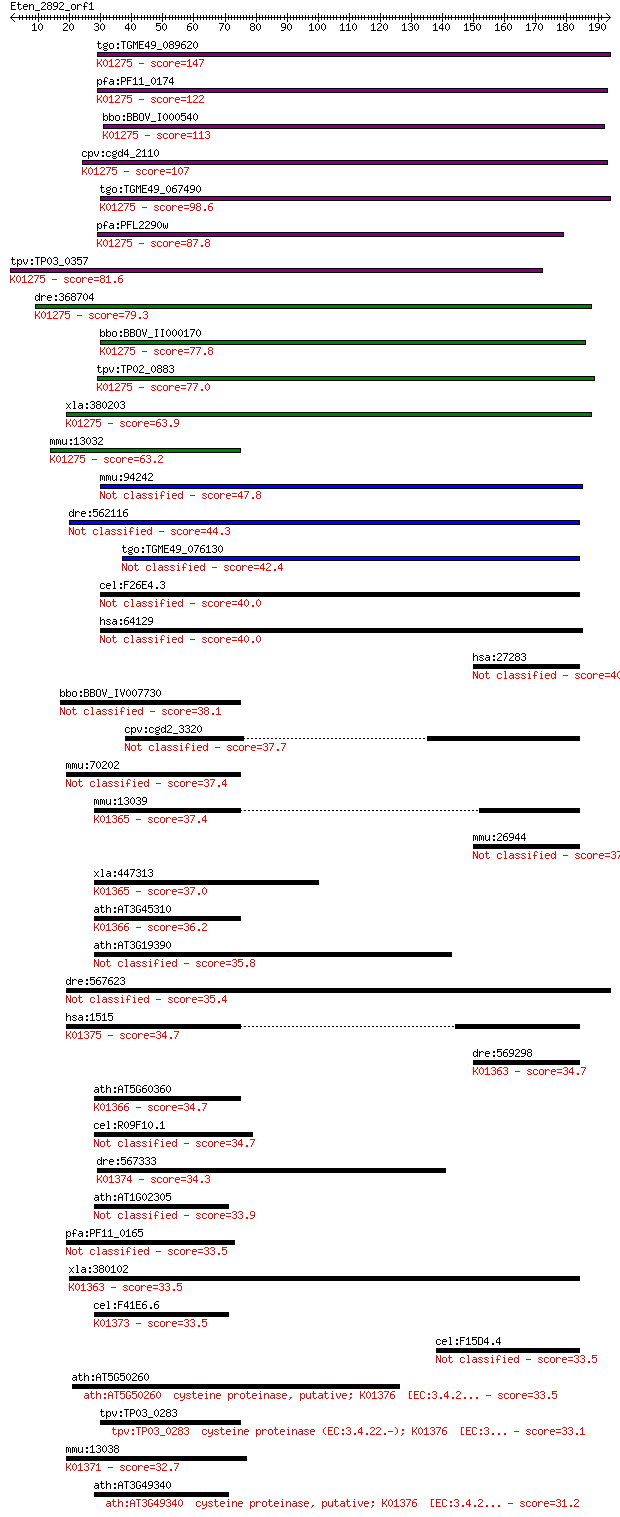
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_089620 cathepsin C (EC:3.4.14.1); K01275 cathepsin ... 147 3e-35
pfa:PF11_0174 cathepsin C, homolog; K01275 cathepsin C [EC:3.4... 122 1e-27
bbo:BBOV_I000540 16.m00694; preprocathepsin c precursor; K0127... 113 5e-25
cpv:cgd4_2110 preprocathepsin c precursor ; K01275 cathepsin C... 107 2e-23
tgo:TGME49_067490 papain family cysteine protease domain-conta... 98.6 1e-20
pfa:PFL2290w preprocathepsin c precursor, putative (EC:3.4.14.... 87.8 2e-17
tpv:TP03_0357 cathepsin C; K01275 cathepsin C [EC:3.4.14.1] 81.6 2e-15
dre:368704 ctsc, cb912, ik:tdsubc_1h2, sb:cb146, wu:fb34g12, w... 79.3 8e-15
bbo:BBOV_II000170 18.m05995; cathepsin C precursor (EC:3.4.22.... 77.8 2e-14
tpv:TP02_0883 cathepsin C; K01275 cathepsin C [EC:3.4.14.1] 77.0 3e-14
xla:380203 ctsc, MGC69126; cathepsin C (EC:3.4.14.1); K01275 c... 63.9 3e-10
mmu:13032 Ctsc, AI047818, DPP1, DPPI; cathepsin C (EC:3.4.14.1... 63.2 5e-10
mmu:94242 Tinagl1, 1110021J17Rik, AZ-1, AZ1, Arg1, Lcn7, TARP,... 47.8 2e-05
dre:562116 tinagl1, si:dkey-158b13.1; tubulointerstitial nephr... 44.3 3e-04
tgo:TGME49_076130 cathepsin C2 (TgCPC2) (EC:3.4.14.1) 42.4 0.001
cel:F26E4.3 hypothetical protein 40.0 0.005
hsa:64129 TINAGL1, ARG1, LCN7, LIECG3, TINAGRP; tubulointersti... 40.0 0.005
hsa:27283 TINAG, TIN-AG; tubulointerstitial nephritis antigen 40.0 0.005
bbo:BBOV_IV007730 23.m06535; cysteine protease 2 38.1
cpv:cgd2_3320 secreted papain like protease, signal peptide 37.7 0.024
mmu:70202 Ctsll3, 2310051M13Rik; cathepsin L-like 3 (EC:3.4.22... 37.4 0.035
mmu:13039 Ctsl, 1190035F06Rik, Ctsl1, MEP, fs, nkt; cathepsin ... 37.4 0.037
mmu:26944 Tinag, AI452335, TIN-ag; tubulointerstitial nephriti... 37.0 0.041
xla:447313 ctsl2, MGC81823; cathepsin L2; K01365 cathepsin L [... 37.0 0.043
ath:AT3G45310 cysteine proteinase, putative; K01366 cathepsin ... 36.2 0.067
ath:AT3G19390 cysteine proteinase, putative / thiol protease, ... 35.8 0.10
dre:567623 zgc:174153 35.4 0.11
hsa:1515 CTSL2, CATL2, CTSU, CTSV, MGC125957; cathepsin L2 (EC... 34.7 0.20
dre:569298 ctsbb; capthepsin B, b; K01363 cathepsin B [EC:3.4.... 34.7 0.21
ath:AT5G60360 AALP; AALP (Arabidopsis aleurain-like protease);... 34.7 0.23
cel:R09F10.1 hypothetical protein 34.7 0.23
dre:567333 ctso; cathepsin O; K01374 cathepsin O [EC:3.4.22.42] 34.3 0.29
ath:AT1G02305 cathepsin B-like cysteine protease, putative 33.9 0.32
pfa:PF11_0165 falcipain-2A 33.5 0.41
xla:380102 cg10992; hypothetical protein MGC52983; K01363 cath... 33.5 0.42
cel:F41E6.6 tag-196; Temporarily Assigned Gene name family mem... 33.5 0.46
cel:F15D4.4 hypothetical protein 33.5 0.47
ath:AT5G50260 cysteine proteinase, putative; K01376 [EC:3.4.2... 33.5 0.50
tpv:TP03_0283 cysteine proteinase (EC:3.4.22.-); K01376 [EC:3... 33.1 0.66
mmu:13038 Ctsk, AI323530, MMS10-Q, Ms10q, catK; cathepsin K (E... 32.7 0.78
ath:AT3G49340 cysteine proteinase, putative; K01376 [EC:3.4.2... 31.2 2.1
> tgo:TGME49_089620 cathepsin C (EC:3.4.14.1); K01275 cathepsin
C [EC:3.4.14.1]
Length=733
Score = 147 bits (370), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 79/165 (47%), Positives = 96/165 (58%), Gaps = 25/165 (15%)
Query 29 KLSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNKNEK 88
+LS QS+LSCSFYNQGC GG PYLVGKHA +IG+ +CM Y A CP
Sbjct 495 ELSAQSILSCSFYNQGCDGGFPYLVGKHARDIGVPQARCMEYQADHTQGCP--------- 545
Query 89 DFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGYVGGCYECLS 148
F K+ +S + + L + G C + RW+AK YGY+GGCYEC
Sbjct 546 -FQKTAS-------------ASESQSMLQADANAGACSE-HARWYAKDYGYIGGCYECNH 590
Query 149 CSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVYTTRGAPHARV 193
CS E++IM EI NGPV A DAPPSLF+Y SGVY + HARV
Sbjct 591 CSGEKQIMLEIYNNGPVPVAFDAPPSLFSYRSGVYDAN-SNHARV 634
> pfa:PF11_0174 cathepsin C, homolog; K01275 cathepsin C [EC:3.4.14.1]
Length=700
Score = 122 bits (305), Expect = 1e-27, Method: Composition-based stats.
Identities = 70/178 (39%), Positives = 100/178 (56%), Gaps = 19/178 (10%)
Query 29 KLSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNKNEK 88
+LS Q+VLSCSFY+QGC+GG PYLV K A GI PY+A + +CP N +K+
Sbjct 430 QLSIQTVLSCSFYDQGCNGGFPYLVSKLAKLQGIPLNVYFPYSATE-ETCP-YNISKHPN 487
Query 89 DFLKSEKDFLKGEINGSGEDSSSNAAF-----------LFSGEATGTCHQG---ENRWFA 134
D S K L+ EIN +++ + + +++ A+ G ENRW+A
Sbjct 488 DMNGSAK--LR-EINAIFNSNNNMSTYNNINNDHHQLGVYANTASSQEQHGISEENRWYA 544
Query 135 KGYGYVGGCYECLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVYTTRGAPHAR 192
K + YVGGCY C C+ E+ +M EI NGP+ ++ +A P + Y+ GVY PHAR
Sbjct 545 KDFNYVGGCYGCNQCNGEKIMMNEIYRNGPIVSSFEASPDFYDYADGVYFVEDFPHAR 602
> bbo:BBOV_I000540 16.m00694; preprocathepsin c precursor; K01275
cathepsin C [EC:3.4.14.1]
Length=546
Score = 113 bits (282), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 67/166 (40%), Positives = 82/166 (49%), Gaps = 53/166 (31%)
Query 31 SPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYT-----AMDLSSCPVLNRNK 85
S Q +L CS +NQGC+GG P+LVGKH TE G+L E PY A+D S VL+ ++
Sbjct 339 SVQDMLECSPFNQGCYGGFPFLVGKHLTEFGVLSEDKSPYRMSNGGAVDTCSVDVLDPSE 398
Query 86 NEKDFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGYVGGCYE 145
RW+A GYGYVGGCYE
Sbjct 399 ---------------------------------------------RWYASGYGYVGGCYE 413
Query 146 CLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVYTTRGAPHA 191
C S E +IM+E+ NGPVA ALDAP SLF YSSG+Y + H
Sbjct 414 CTS---ELEIMREVYHNGPVAVALDAPQSLFQYSSGIYDDNPSNHG 456
> cpv:cgd4_2110 preprocathepsin c precursor ; K01275 cathepsin
C [EC:3.4.14.1]
Length=635
Score = 107 bits (268), Expect = 2e-23, Method: Composition-based stats.
Identities = 65/169 (38%), Positives = 84/169 (49%), Gaps = 35/169 (20%)
Query 24 QEAAVKLSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNR 83
+E + LSPQSVLSCS +NQGC GG P+LVG+ A EIGI E+CM Y A C
Sbjct 385 REEKILLSPQSVLSCSPFNQGCEGGYPFLVGRQAEEIGISSEKCMGYYADSNQEC----- 439
Query 84 NKNEKDFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGYVGGC 143
+ F+ EI E C +GE R +A+ YGYVGGC
Sbjct 440 ---------NFSPFITPEI-----------------EDRIYCEEGE-RMYAEEYGYVGGC 472
Query 144 YECLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVYTTRGAPHAR 192
Y C E ++ +EI NGP+A A+ SL Y +GVY + H +
Sbjct 473 Y---GCCDEDRMKEEIFKNGPIAVAMHIDTSLLVYDNGVYDSIPNDHTK 518
> tgo:TGME49_067490 papain family cysteine protease domain-containing
protein (EC:3.4.14.1); K01275 cathepsin C [EC:3.4.14.1]
Length=622
Score = 98.6 bits (244), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 61/164 (37%), Positives = 89/164 (54%), Gaps = 36/164 (21%)
Query 30 LSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNKNEKD 89
LS QS+LSCS YNQGC GG P+LVGKHA E G EQC +S+C + +
Sbjct 400 LSAQSILSCSPYNQGCDGGYPFLVGKHAKEFGFGTEQC------QVSACKMFH------- 446
Query 90 FLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGYVGGCYECLSC 149
+++ +IN E +F + +C +AK Y YVGG YE C
Sbjct 447 -------YIQHKINSVAE--------IFCQPRSPSCFL-----YAKDYNYVGGFYE--GC 484
Query 150 SAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVYTTRGAPHARV 193
+ E+K+M E+ +GPV A+DAP +LF Y SG++ ++ + H ++
Sbjct 485 N-EEKMMNEMYHHGPVVVAIDAPDTLFMYQSGLFDSQPSEHGKI 527
> pfa:PFL2290w preprocathepsin c precursor, putative (EC:3.4.14.1);
K01275 cathepsin C [EC:3.4.14.1]
Length=590
Score = 87.8 bits (216), Expect = 2e-17, Method: Composition-based stats.
Identities = 54/150 (36%), Positives = 74/150 (49%), Gaps = 36/150 (24%)
Query 29 KLSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNKNEK 88
+LS QS+LSCS YNQGC GG P+LVGKH E GI+ EQ M Y D ++C + N N
Sbjct 351 RLSHQSILSCSPYNQGCDGGYPFLVGKHMYEYGIIPEQYMHYENNDYNNCIMDMGNYNH- 409
Query 89 DFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGYVGGCYECLS 148
L + +K EI ++ Y Y+ GCYE
Sbjct 410 --LNKQNRNIK-EI-----------------------------YYVSDYNYINGCYE--- 434
Query 149 CSAEQKIMKEIMTNGPVAAALDAPPSLFAY 178
C+ E ++M EI+ NGP+ AA++A L +
Sbjct 435 CTNEYEMMNEIILNGPIVAAINATSELLNF 464
> tpv:TP03_0357 cathepsin C; K01275 cathepsin C [EC:3.4.14.1]
Length=501
Score = 81.6 bits (200), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 55/174 (31%), Positives = 73/174 (41%), Gaps = 52/174 (29%)
Query 1 RLQQQQQEGEEQQQQQLQQLQQ---QQEAAVKLSPQSVLSCSFYNQGCHGGLPYLVGKHA 57
R+++ ++ E + +L L++ + + LS LSC YNQGC GG P VGK A
Sbjct 260 RIRELLKKPEYKSDARLLHLEKVLSDKNFNINLS----LSCIPYNQGCKGGFPVNVGKFA 315
Query 58 TEIGILDEQCMPYTAMDLSSCPVLNRNKNEKDFLKSEKDFLKGEINGSGEDSSSNAAFLF 117
E G++ + P +D CP E++F
Sbjct 316 EEFGLILDNEKPEEVVDNLKCP------------PKEENF-------------------- 343
Query 118 SGEATGTCHQGENRWFAKGYGYVGGCYECLSCSAEQKIMKEIMTNGPVAAALDA 171
R FA GYVGGCYEC CS E IM EIM NGPV A +D
Sbjct 344 -------------RLFASNVGYVGGCYECTRCSGETLIMNEIMLNGPVVAGIDG 384
> dre:368704 ctsc, cb912, ik:tdsubc_1h2, sb:cb146, wu:fb34g12,
wu:fj58d01; cathepsin C (EC:3.4.14.1); K01275 cathepsin C [EC:3.4.14.1]
Length=455
Score = 79.3 bits (194), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 56/179 (31%), Positives = 76/179 (42%), Gaps = 52/179 (29%)
Query 9 GEEQQQQQLQQLQQQQEAAVKLSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCM 68
G + + ++Q QQ SPQ V+SCS Y+QGC GG PYL+GK+ + GI++E C
Sbjct 258 GMLEARVRIQTNNTQQPV---FSPQQVVSCSQYSQGCDGGFPYLIGKYIQDFGIVEEDCF 314
Query 69 PYTAMDLSSCPVLNRNKNEKDFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQG 128
PYT G DS N C
Sbjct 315 PYT----------------------------------GSDSPCNLP--------AKC--- 329
Query 129 ENRWFAKGYGYVGGCYECLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVYTTRG 187
+++A Y YVGG Y CS E +M E++ NGP+ AL+ P Y G+Y G
Sbjct 330 -TKYYASDYHYVGGFYG--GCS-ESAMMLELVKNGPMGVALEVYPDFMNYKEGIYHHTG 384
> bbo:BBOV_II000170 18.m05995; cathepsin C precursor (EC:3.4.22.-);
K01275 cathepsin C [EC:3.4.14.1]
Length=530
Score = 77.8 bits (190), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 47/157 (29%), Positives = 65/157 (41%), Gaps = 53/157 (33%)
Query 30 LSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNKNEK- 88
P+ V CS YNQGC GG PYL+GK E GIL +N ++
Sbjct 324 FDPKDVTDCSMYNQGCDGGYPYLMGKQMREFGILT-----------------TKNAGQQC 366
Query 89 DFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGYVGGCYECLS 148
L +E+ + FA+ YGYVGGC++C +
Sbjct 367 TLLSTERRY-----------------------------------FARDYGYVGGCHQCTA 391
Query 149 CSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVYTT 185
C + IM+EI+ NGPV A+DA Y + T+
Sbjct 392 CQGDALIMREILANGPVVTAIDAAVLTADYDGHIITS 428
> tpv:TP02_0883 cathepsin C; K01275 cathepsin C [EC:3.4.14.1]
Length=365
Score = 77.0 bits (188), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 53/160 (33%), Positives = 70/160 (43%), Gaps = 51/160 (31%)
Query 29 KLSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNKNEK 88
KLS +LS S Y+QGC GG LVGKH E+GI + + +
Sbjct 181 KLSIHDLLS-SAYSQGCFGGFLMLVGKHIKELGI---------------------HSDSE 218
Query 89 DFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGYVGGCYECLS 148
FL + + ED + +W+ YGYVGGCYEC
Sbjct 219 TFLNTLRTL---------EDIKFDM-----------------KWYIDSYGYVGGCYEC-- 250
Query 149 CSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVYTTRGA 188
+ E +M EI+TNGP+A A+ +PP LF Y G T A
Sbjct 251 -TNEMNMMNEIITNGPIAVAIYSPPQLFYYKHGWEYTNHA 289
> xla:380203 ctsc, MGC69126; cathepsin C (EC:3.4.14.1); K01275
cathepsin C [EC:3.4.14.1]
Length=458
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 50/170 (29%), Positives = 67/170 (39%), Gaps = 50/170 (29%)
Query 19 QLQQQQEAAVKLSPQSVLSCSFYNQGCHGGLPYLV-GKHATEIGILDEQCMPYTAMDLSS 77
Q+Q Q LSPQ V+SCS Y+QGC GG PYL+ GK+ + GI++E PY D S
Sbjct 267 QIQSQLSQKPILSPQQVVSCSNYSQGCDGGFPYLIAGKYLNDFGIVEESDFPYIGSD-SP 325
Query 78 CPVLNRNKNEKDFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGY 137
C T R++ Y
Sbjct 326 C---------------------------------------------TLKDSYQRYYTAEY 340
Query 138 GYVGGCYECLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVYTTRG 187
YVGG Y C+ E + E++ GP++ A + Y SGVY G
Sbjct 341 HYVGGFYG--GCN-EAYMKLELVLGGPLSVAFEVYDDFIHYRSGVYHHTG 387
> mmu:13032 Ctsc, AI047818, DPP1, DPPI; cathepsin C (EC:3.4.14.1);
K01275 cathepsin C [EC:3.4.14.1]
Length=462
Score = 63.2 bits (152), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 29/62 (46%), Positives = 42/62 (67%), Gaps = 2/62 (3%)
Query 14 QQQLQQLQQQQEAAVKLSPQSVLSCSFYNQGCHGGLPYLV-GKHATEIGILDEQCMPYTA 72
+ +++ L + + LSPQ V+SCS Y QGC GG PYL+ GK+A + G+++E C PYTA
Sbjct 267 EARIRILTNNSQTPI-LSPQEVVSCSPYAQGCDGGFPYLIAGKYAQDFGVVEESCFPYTA 325
Query 73 MD 74
D
Sbjct 326 KD 327
> mmu:94242 Tinagl1, 1110021J17Rik, AZ-1, AZ1, Arg1, Lcn7, TARP,
Tinagl; tubulointerstitial nephritis antigen-like 1
Length=466
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 43/159 (27%), Positives = 67/159 (42%), Gaps = 34/159 (21%)
Query 30 LSPQSVLSC-SFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNKNEK 88
LSPQ++LSC + + QGC GG G++ + C P++ R +NE
Sbjct 253 LSPQNLLSCDTHHQQGCRGGRLDGAWWFLRRRGVVSDNCYPFSG----------REQNEA 302
Query 89 DFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGE---NRWFAKGYGYVGGCYE 145
+ + + G G+ +AT C G+ N + Y G
Sbjct 303 S--PTPRCMMHSRAMGRGKR-----------QATSRCPNGQVDSNDIYQVTPAYRLG--- 346
Query 146 CLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVYT 184
S E++IMKE+M NGPV A ++ F Y G+Y+
Sbjct 347 ----SDEKEIMKELMENGPVQALMEVHEDFFLYQRGIYS 381
> dre:562116 tinagl1, si:dkey-158b13.1; tubulointerstitial nephritis
antigen-like 1
Length=471
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 40/169 (23%), Positives = 66/169 (39%), Gaps = 37/169 (21%)
Query 20 LQQQQEAAVKLSPQSVLSCSFYNQ-GCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSC 78
+Q +LSPQ+++SC +Q GC GG G++ + C P++ + S+
Sbjct 241 IQSMGHMTPQLSPQNLISCDTRHQDGCAGGRIDGAWWFMRRRGVVTQDCYPFSPPEQSAV 300
Query 79 PVLNRNKNEKDFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYG 138
V + ++ G G+ +AT C +
Sbjct 301 EV-------------ARCMMQSRAVGRGKR-----------QATAHC--------PNSHS 328
Query 139 YVGGCYECLS----CSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY 183
Y Y+ + E +IMKEIM NGPV A ++ F Y SG++
Sbjct 329 YHNDIYQSTPPYRLSTNENEIMKEIMDNGPVQAIMEVHEDFFVYKSGIF 377
> tgo:TGME49_076130 cathepsin C2 (TgCPC2) (EC:3.4.14.1)
Length=753
Score = 42.4 bits (98), Expect = 0.001, Method: Composition-based stats.
Identities = 42/148 (28%), Positives = 65/148 (43%), Gaps = 17/148 (11%)
Query 37 SCSFYNQGCHGGLPYLVGKHATEIGILDEQCMP-YTAMDLSSCPVLNRNKNEKDFLKSEK 95
SC+ YNQGC GG +L K E G E C+ Y AM L+ N L++
Sbjct 425 SCNVYNQGCGGGYVFLALKFGQEHGFRTEDCVSEYHAMADKHKGSLSPN------LQTCF 478
Query 96 DFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGYVGGCYECLSCSAEQKI 155
D L G++ S + A +C+ + YVGG Y CS E +
Sbjct 479 D-LGGQLGTSAFGCQAPPA---RASLPDSCNLSVK---VTSWHYVGGVYG--GCS-EDDM 528
Query 156 MKEIMTNGPVAAALDAPPSLFAYSSGVY 183
++ + +GP+AA+++ + Y GV+
Sbjct 529 LRTLWEHGPMAASIEPTIAFTVYKKGVF 556
> cel:F26E4.3 hypothetical protein
Length=452
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 37/155 (23%), Positives = 59/155 (38%), Gaps = 35/155 (22%)
Query 30 LSPQSVLSCSFYNQ-GCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNKNEK 88
LS Q +LSC+ + Q GC GG + ++G++ + C PY V +++
Sbjct 235 LSSQQLLSCNQHRQKGCEGGYLDRAWWYIRKLGVVGDHCYPY---------VSGQSREPG 285
Query 89 DFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGYVGGCYECLS 148
L ++D+ + S + AF + +
Sbjct 286 HCLIPKRDYTNRQGLRCPSGSQDSTAFKMTPPYKVS------------------------ 321
Query 149 CSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY 183
S E+ I E+MTNGPV A F Y+ GVY
Sbjct 322 -SREEDIQTELMTNGPVQATFVVHEDFFMYAGGVY 355
> hsa:64129 TINAGL1, ARG1, LCN7, LIECG3, TINAGRP; tubulointerstitial
nephritis antigen-like 1
Length=436
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 40/159 (25%), Positives = 64/159 (40%), Gaps = 34/159 (21%)
Query 30 LSPQSVLSCSFYNQ-GCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNKNEK 88
LSPQ++LSC + Q GC GG G++ + C P++ R ++E
Sbjct 223 LSPQNLLSCDTHQQQGCRGGRLDGAWWFLRRRGVVSDHCYPFSG----------RERDEA 272
Query 89 DFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTC---HQGENRWFAKGYGYVGGCYE 145
+ + G G+ +AT C + N + Y G
Sbjct 273 G--PAPPCMMHSRAMGRGKR-----------QATAHCPNSYVNNNDIYQVTPVYRLG--- 316
Query 146 CLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVYT 184
S +++IMKE+M NGPV A ++ F Y G+Y+
Sbjct 317 ----SNDKEIMKELMENGPVQALMEVHEDFFLYKGGIYS 351
> hsa:27283 TINAG, TIN-AG; tubulointerstitial nephritis antigen
Length=476
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 17/34 (50%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 150 SAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY 183
S E +IMKEIM NGPV A + F Y +G+Y
Sbjct 359 SNETEIMKEIMQNGPVQAIMQVREDFFHYKTGIY 392
> bbo:BBOV_IV007730 23.m06535; cysteine protease 2
Length=445
Score = 38.1 bits (87), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 31/58 (53%), Gaps = 0/58 (0%)
Query 17 LQQLQQQQEAAVKLSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMD 74
++ L ++Q+ V+LS Q ++SC NQGC+GG + GI + PY A D
Sbjct 269 VESLLKRQKTDVRLSEQELVSCQLGNQGCNGGYSDYALNYIKFNGIHRSEEWPYLAAD 326
> cpv:cgd2_3320 secreted papain like protease, signal peptide
Length=819
Score = 37.7 bits (86), Expect = 0.024, Method: Composition-based stats.
Identities = 17/38 (44%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 38 CSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDL 75
C+ YNQGC GGL L K A E+G+ + C+ A +L
Sbjct 437 CNVYNQGCGGGLITLAFKFAQEVGVRTQDCVDDYAKNL 474
Score = 29.3 bits (64), Expect = 8.0, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 24/49 (48%), Gaps = 3/49 (6%)
Query 135 KGYGYVGGCYECLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY 183
K Y YV Y + + IM+ + GPVA +L+ Y+SGV+
Sbjct 669 KEYSYVNNVY---GKTTARDIMESLWNEGPVAVSLEPTLEFSLYNSGVF 714
> mmu:70202 Ctsll3, 2310051M13Rik; cathepsin L-like 3 (EC:3.4.22.15)
Length=331
Score = 37.4 bits (85), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 34/59 (57%), Gaps = 3/59 (5%)
Query 19 QLQQQQEAAVKLSPQSVLSCSFY--NQGCHGGLPYLVGKHATEIGILDEQC-MPYTAMD 74
Q+ ++ V LS Q+++ CS+ NQGC GGLP L ++ + G LD PY A++
Sbjct 151 QMFRKTGKLVPLSVQNLVDCSWSQGNQGCDGGLPDLAFQYVKDNGGLDTSVSYPYEALN 209
> mmu:13039 Ctsl, 1190035F06Rik, Ctsl1, MEP, fs, nkt; cathepsin
L (EC:3.4.22.15); K01365 cathepsin L [EC:3.4.22.15]
Length=334
Score = 37.4 bits (85), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 17/33 (51%), Positives = 24/33 (72%), Gaps = 1/33 (3%)
Query 152 EQKIMKEIMTNGPVAAALDAP-PSLFAYSSGVY 183
E+ +MK + T GP++ A+DA PSL YSSG+Y
Sbjct 232 EKALMKAVATVGPISVAMDASHPSLQFYSSGIY 264
Score = 34.7 bits (78), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 21/50 (42%), Positives = 29/50 (58%), Gaps = 3/50 (6%)
Query 28 VKLSPQSVLSCSFY--NQGCHGGLPYLVGKHATEIGILD-EQCMPYTAMD 74
+ LS Q+++ CS NQGC+GGL ++ E G LD E+ PY A D
Sbjct 159 ISLSEQNLVDCSHAQGNQGCNGGLMDFAFQYIKENGGLDSEESYPYEAKD 208
> mmu:26944 Tinag, AI452335, TIN-ag; tubulointerstitial nephritis
antigen
Length=475
Score = 37.0 bits (84), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 150 SAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY 183
S E +IM+EI+ NGPV A + F Y +G+Y
Sbjct 358 SNETEIMREIIQNGPVQAIMQVHEDFFYYKTGIY 391
> xla:447313 ctsl2, MGC81823; cathepsin L2; K01365 cathepsin L
[EC:3.4.22.15]
Length=335
Score = 37.0 bits (84), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 30/82 (36%), Positives = 43/82 (52%), Gaps = 11/82 (13%)
Query 28 VKLSPQSVLSCSFY--NQGCHGGLPYLVGKHATEIGILD-EQCMPYTAMDLSSC---PVL 81
+ LS Q+++ CS NQGC+GGL ++ + G +D E PYTA D C P
Sbjct 159 ISLSEQNLVDCSRAQGNQGCNGGLMDQAFQYVKDNGGIDSEDSYPYTAKDDQECHYDPNY 218
Query 82 NRNKNEKDFLK----SEKDFLK 99
N + N+ F+ SEKD +K
Sbjct 219 N-SANDTGFVDVPSGSEKDLMK 239
> ath:AT3G45310 cysteine proteinase, putative; K01366 cathepsin
H [EC:3.4.22.16]
Length=357
Score = 36.2 bits (82), Expect = 0.067, Method: Compositional matrix adjust.
Identities = 21/50 (42%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query 28 VKLSPQSVLSC--SFYNQGCHGGLPYLVGKHATEIGILD-EQCMPYTAMD 74
+ LS Q ++ C +F N GCHGGLP ++ G LD E+ PYT D
Sbjct 186 ISLSEQQLVDCAGTFNNFGCHGGLPSQAFEYIKYNGGLDTEEAYPYTGKD 235
> ath:AT3G19390 cysteine proteinase, putative / thiol protease,
putative
Length=452
Score = 35.8 bits (81), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 40/131 (30%), Positives = 56/131 (42%), Gaps = 18/131 (13%)
Query 28 VKLSPQSVLSC-SFYNQGCHGGLPYLVGKHATEIGILD-EQCMPYTAMDLSSCPVLNRN- 84
+ LS Q ++ C + YN GC GGL K E G +D E+ PY A D++ C +N
Sbjct 174 ISLSEQELVDCDTSYNDGCGGGLMDYAFKFIIENGGIDTEEDYPYIATDVNVCNSDKKNT 233
Query 85 ------------KNEKDFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQG-ENR 131
+N++ LK K I+ + E SG TGTC ++
Sbjct 234 RVVTIDGYEDVPQNDEKSLK--KALANQPISVAIEAGGRAFQLYTSGVFTGTCGTSLDHG 291
Query 132 WFAKGYGYVGG 142
A GYG GG
Sbjct 292 VVAVGYGSEGG 302
> dre:567623 zgc:174153
Length=336
Score = 35.4 bits (80), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 46/182 (25%), Positives = 65/182 (35%), Gaps = 62/182 (34%)
Query 19 QLQQQQEAAVKLSPQSVLSCSFY--NQGCHGGLPYLVGKHATEIGILD-EQCMPYTAMDL 75
QL ++ + +S Q+++ CS NQGC+GGL L ++ E LD EQ PY A D
Sbjct 151 QLFRKTGKLISMSEQNLVDCSRPQGNQGCNGGLMDLAFQYVKENKGLDSEQSYPYLARDD 210
Query 76 SSC---PVLNRNKNEKDFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRW 132
C P N
Sbjct 211 LPCRYDPRFN-------------------------------------------------- 220
Query 133 FAKGYGYVGGCYECLSCSAEQKIMKEIMTNGPVAAALDAPP-SLFAYSSGVYTTRGAPHA 191
AK G+V + E +M + GPV+ A+DA SL Y SG+Y R +
Sbjct 221 VAKSTGFVD-----IPSGNEPALMNAVAAVGPVSVAIDASHQSLQFYQSGIYYERACSSS 275
Query 192 RV 193
R+
Sbjct 276 RL 277
> hsa:1515 CTSL2, CATL2, CTSU, CTSV, MGC125957; cathepsin L2 (EC:3.4.22.43);
K01375 cathepsin V [EC:3.4.22.43]
Length=334
Score = 34.7 bits (78), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 33/59 (55%), Gaps = 3/59 (5%)
Query 19 QLQQQQEAAVKLSPQSVLSCSFY--NQGCHGGLPYLVGKHATEIGILD-EQCMPYTAMD 74
Q+ ++ V LS Q+++ CS NQGC+GG ++ E G LD E+ PY A+D
Sbjct 150 QMFRKTGKLVSLSEQNLVDCSRPQGNQGCNGGFMARAFQYVKENGGLDSEESYPYVAVD 208
Score = 32.7 bits (73), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Query 144 YECLSCSAEQKIMKEIMTNGPVAAALDAPPSLF-AYSSGVY 183
+ ++ E+ +MK + T GP++ A+DA S F Y SG+Y
Sbjct 225 FTVVAPGKEKALMKAVATVGPISVAMDAGHSSFQFYKSGIY 265
> dre:569298 ctsbb; capthepsin B, b; K01363 cathepsin B [EC:3.4.22.1]
Length=326
Score = 34.7 bits (78), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 17/34 (50%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 150 SAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY 183
S +Q+IM E+ TNGPV AA Y SGVY
Sbjct 228 SDQQQIMTELYTNGPVEAAFTVYEDFPLYKSGVY 261
> ath:AT5G60360 AALP; AALP (Arabidopsis aleurain-like protease);
cysteine-type peptidase; K01366 cathepsin H [EC:3.4.22.16]
Length=357
Score = 34.7 bits (78), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query 28 VKLSPQSVLSCS--FYNQGCHGGLPYLVGKHATEIGILD-EQCMPYTAMD 74
+ LS Q ++ C+ F N GC+GGLP ++ G LD E+ PYT D
Sbjct 186 ISLSEQQLVDCAGAFNNYGCNGGLPSQAFEYIKSNGGLDTEKAYPYTGKD 235
> cel:R09F10.1 hypothetical protein
Length=383
Score = 34.7 bits (78), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 24/51 (47%), Gaps = 0/51 (0%)
Query 28 VKLSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSC 78
V LS Q ++ C N GC GG K E G+ E+ PY+A+ C
Sbjct 213 VSLSEQEMVDCDGRNNGCSGGYRPYAMKFVKENGLESEKEYPYSALKHDQC 263
> dre:567333 ctso; cathepsin O; K01374 cathepsin O [EC:3.4.22.42]
Length=334
Score = 34.3 bits (77), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 38/150 (25%), Positives = 56/150 (37%), Gaps = 38/150 (25%)
Query 29 KLSPQSVLSCSFYNQGCHGGLPY--LVGKHATEIGILDEQCMPYTAMD-------LSSCP 79
+LS Q V+ CS+ NQGC+GG P L +++ ++ E P+ D +
Sbjct 167 QLSVQQVIDCSYQNQGCNGGSPVEALYWLTQSKLKLVSEAEYPFKGADGVCQFFPQAHAG 226
Query 80 VLNRNKNEKDFLKSE----------------------KDFLKGEINGSGEDSSSNAAFLF 117
V RN + DF E +D+L G I +N A L
Sbjct 227 VAVRNYSAYDFSGQEEVMMSALVDFGPLVVIVDAISWQDYLGGIIQHHCSSHKANHAVLI 286
Query 118 SG-EATGTCHQGENR------WFAKGYGYV 140
+G + TG R W GY Y+
Sbjct 287 TGYDTTGEVPYWIVRNSWGTSWGDDGYAYI 316
> ath:AT1G02305 cathepsin B-like cysteine protease, putative
Length=362
Score = 33.9 bits (76), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 25/45 (55%), Gaps = 2/45 (4%)
Query 28 VKLSPQSVLSCSFY--NQGCHGGLPYLVGKHATEIGILDEQCMPY 70
V LS +L+C + QGC+GG P ++ G++ E+C PY
Sbjct 155 VSLSVNDLLACCGFLCGQGCNGGYPIAAWRYFKHHGVVTEECDPY 199
> pfa:PF11_0165 falcipain-2A
Length=484
Score = 33.5 bits (75), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query 19 QLQQQQEAAVKLSPQSVLSCSFYNQGCHGGLPYLVGKHATEI-GILDEQCMPYTA 72
Q ++ + LS Q ++ CSF N GC+GGL + E+ GI + PY +
Sbjct 297 QYAIRKNKLITLSEQELVDCSFKNYGCNGGLINNAFEDMIELGGICTDDDYPYVS 351
> xla:380102 cg10992; hypothetical protein MGC52983; K01363 cathepsin
B [EC:3.4.22.1]
Length=333
Score = 33.5 bits (75), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 42/178 (23%), Positives = 66/178 (37%), Gaps = 46/178 (25%)
Query 20 LQQQQEAAVKLSPQSVLSCSFYN--QGCHGGLPYLVGKHATEIGIL-----DEQ--CMPY 70
+ + V++S + +LSC + GC+GG P + TE G++ D C PY
Sbjct 123 VHTNGKVNVEVSAEDLLSCCGFKCGMGCNGGYPSGAWRFWTETGLVSGGLYDSHVGCRPY 182
Query 71 TAMDLSSCPVLNRNKNEKDFLKSEKDFLKGEINGS-----GEDSSSNAAFLFSGEATGTC 125
+ + C + +NGS GE+ + E
Sbjct 183 S---IPPC--------------------EHHVNGSRPSCKGEEGDTPKCMKTCEEGYTPA 219
Query 126 HQGENRWFAKGYGYVGGCYECLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY 183
+ + + A YG S+E++IM +I NGPV A Y SGVY
Sbjct 220 YGSDKHFGATSYGVP---------SSEKEIMADIYKNGPVEGAFVVYADFPLYKSGVY 268
> cel:F41E6.6 tag-196; Temporarily Assigned Gene name family member
(tag-196); K01373 cathepsin F [EC:3.4.22.41]
Length=477
Score = 33.5 bits (75), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 24/44 (54%), Gaps = 1/44 (2%)
Query 28 VKLSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILD-EQCMPY 70
V LS Q ++ C +QGC+GGLP K +G L+ E PY
Sbjct 309 VSLSEQELVDCDSMDQGCNGGLPSNAYKEIIRMGGLEPEDAYPY 352
> cel:F15D4.4 hypothetical protein
Length=608
Score = 33.5 bits (75), Expect = 0.47, Method: Composition-based stats.
Identities = 13/46 (28%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 138 GYVGGCYECLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY 183
GY+ G + ++ +++ + GP+A + A P ++ YS GVY
Sbjct 341 GYISGNFTAAQLITMEQNIEDKVRKGPIAVGMAAGPDIYKYSEGVY 386
> ath:AT5G50260 cysteine proteinase, putative; K01376 [EC:3.4.22.-]
Length=361
Score = 33.5 bits (75), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 34/117 (29%), Positives = 52/117 (44%), Gaps = 12/117 (10%)
Query 21 QQQQEAAVKLSPQSVLSC-SFYNQGCHGGLPYLVGKHATEI-GILDEQCMPYTAMDLS-- 76
Q + + LS Q ++ C + NQGC+GGL L + E G+ E PY A D +
Sbjct 164 QIRTKKLTSLSEQELVDCDTNQNQGCNGGLMDLAFEFIKEKGGLTSELVYPYKASDETCD 223
Query 77 ----SCPVLNRNKNEKDFLKSEKDFLKGEIN----GSGEDSSSNAAFLFSGEATGTC 125
+ PV++ + +E SE D +K N + + S+ F G TG C
Sbjct 224 TNKENAPVVSIDGHEDVPKNSEDDLMKAVANQPVSVAIDAGGSDFQFYSEGVFTGRC 280
> tpv:TP03_0283 cysteine proteinase (EC:3.4.22.-); K01376 [EC:3.4.22.-]
Length=441
Score = 33.1 bits (74), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 30 LSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMD 74
LS Q +++C + GC GGLP ++ GI E +PY +D
Sbjct 275 LSEQELVNCDKSSMGCSGGLPITAMEYIHSKGISFESEIPYIGID 319
> mmu:13038 Ctsk, AI323530, MMS10-Q, Ms10q, catK; cathepsin K
(EC:3.4.22.38); K01371 cathepsin K [EC:3.4.22.38]
Length=329
Score = 32.7 bits (73), Expect = 0.78, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 1/59 (1%)
Query 19 QLQQQQEAAVKLSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILD-EQCMPYTAMDLS 76
QL+++ + LSPQ+++ C N GC GG ++ + G +D E PY D S
Sbjct 151 QLKKKTGKLLALSPQNLVDCVTENYGCGGGYMTTAFQYVQQNGGIDSEDAYPYVGQDES 209
> ath:AT3G49340 cysteine proteinase, putative; K01376 [EC:3.4.22.-]
Length=341
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 22/44 (50%), Gaps = 1/44 (2%)
Query 28 VKLSPQSVLSCSFYNQGCHGGLPYLVGKHATEI-GILDEQCMPY 70
V LS Q +L CS N GC GG+ + + E GI E PY
Sbjct 172 VSLSEQQLLDCSTENNGCGGGIMWKAFDYIKENQGITTEDNYPY 215
Lambda K H
0.314 0.130 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5623228644
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40