bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2929_orf1
Length=135
Score E
Sequences producing significant alignments: (Bits) Value
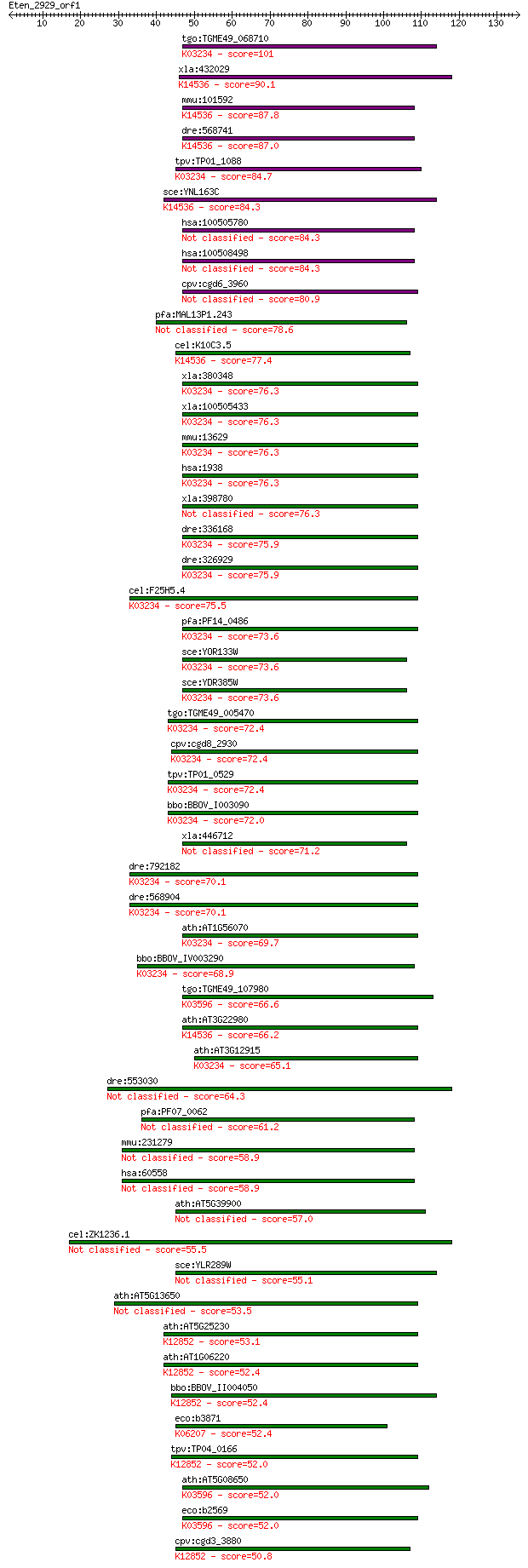
tgo:TGME49_068710 elongation factor Tu GTP-binding domain-cont... 101 8e-22
xla:432029 eftud1, MGC83880; elongation factor Tu GTP binding ... 90.1 2e-18
mmu:101592 Eftud1, 4932434J20Rik, 6030468D11Rik, AI451340, AU0... 87.8 9e-18
dre:568741 Elongation FacTor family member (eft-2)-like; K1453... 87.0 2e-17
tpv:TP01_1088 elongation factor Tu; K03234 elongation factor 2 84.7
sce:YNL163C RIA1, EFL1; Cytoplasmic GTPase involved in biogene... 84.3 8e-17
hsa:100505780 hypothetical protein LOC100505780 84.3 1e-16
hsa:100508498 hypothetical protein LOC100508498 84.3 1e-16
cpv:cgd6_3960 elongation factor-like protein 80.9 1e-15
pfa:MAL13P1.243 elongation factor Tu, putative 78.6 5e-15
cel:K10C3.5 hypothetical protein; K14536 ribosome assembly pro... 77.4 1e-14
xla:380348 eef2.1, MGC53560, eef-2, eef2, ef2; eukaryotic tran... 76.3 3e-14
xla:100505433 hypothetical protein LOC100505433; K03234 elonga... 76.3 3e-14
mmu:13629 Eef2, Ef-2, MGC98463; eukaryotic translation elongat... 76.3 3e-14
hsa:1938 EEF2, EEF-2, EF2; eukaryotic translation elongation f... 76.3 3e-14
xla:398780 hypothetical protein MGC68699 76.3 3e-14
dre:336168 eef2l2, fj53d02, si:ch211-113n10.4, wu:fj53d02; euk... 75.9 3e-14
dre:326929 eef2b, EEF2, MGC63584, eef2l, fe49h02, wu:fe49h02, ... 75.9 4e-14
cel:F25H5.4 eft-2; Elongation FacTor family member (eft-2); K0... 75.5 4e-14
pfa:PF14_0486 elongation factor 2; K03234 elongation factor 2 73.6
sce:YOR133W EFT1; Eft1p; K03234 elongation factor 2 73.6
sce:YDR385W EFT2; Eft2p; K03234 elongation factor 2 73.6
tgo:TGME49_005470 elongation factor 2, putative ; K03234 elong... 72.4 3e-13
cpv:cgd8_2930 Eft2p GTpase; translation elongation factor 2 (E... 72.4 4e-13
tpv:TP01_0529 elongation factor 2; K03234 elongation factor 2 72.4
bbo:BBOV_I003090 19.m02240; elongation factor 2, EF-2; K03234 ... 72.0 4e-13
xla:446712 eef2.2, MGC84492, eef-2, ef2, eft-2; eukaryotic tra... 71.2 8e-13
dre:792182 eef2a.1, MGC113191, si:dkey-110c1.3, zgc:113191; eu... 70.1 2e-12
dre:568904 eef2a.2, si:dkey-110c1.4; eukaryotic translation el... 70.1 2e-12
ath:AT1G56070 LOS1; LOS1; copper ion binding / translation elo... 69.7 3e-12
bbo:BBOV_IV003290 21.m02927; Elongation factor Tu-like protein... 68.9 4e-12
tgo:TGME49_107980 GTP-binding protein lepA, putative (EC:2.7.7... 66.6 2e-11
ath:AT3G22980 elongation factor Tu family protein; K14536 ribo... 66.2 2e-11
ath:AT3G12915 GTP binding / GTPase; K03234 elongation factor 2 65.1
dre:553030 guf1, im:6906935, wu:fc88c11; GUF1 GTPase homolog (... 64.3 1e-10
pfa:PF07_0062 GTP-binding translation elongation factor tu fam... 61.2 9e-10
mmu:231279 Guf1, 4631409J12, AA407526, EF-4; GUF1 GTPase homol... 58.9 4e-09
hsa:60558 GUF1, EF-4, FLJ13220; GUF1 GTPase homolog (S. cerevi... 58.9 4e-09
ath:AT5G39900 GTP binding / GTPase/ translation elongation factor 57.0 2e-08
cel:ZK1236.1 hypothetical protein 55.5 4e-08
sce:YLR289W GUF1; Guf1p 55.1 7e-08
ath:AT5G13650 elongation factor family protein 53.5 2e-07
ath:AT5G25230 elongation factor Tu family protein; K12852 116 ... 53.1 2e-07
ath:AT1G06220 MEE5; MEE5 (MATERNAL EFFECT EMBRYO ARREST 5); GT... 52.4 4e-07
bbo:BBOV_II004050 18.m06335; u5 small nuclear ribonuclear prot... 52.4 4e-07
eco:b3871 typA, bipA, ECK3864, JW5571, yihK; GTP-binding prote... 52.4 4e-07
tpv:TP04_0166 U5 small nuclear ribonucleoprotein; K12852 116 k... 52.0 5e-07
ath:AT5G08650 GTP-binding protein LepA, putative; K03596 GTP-b... 52.0 5e-07
eco:b2569 lepA, ECK2567, JW2553; back-translocating elongation... 52.0 6e-07
cpv:cgd3_3880 Snu114p GTpase, U5 snRNP-specific protein, 116 k... 50.8 1e-06
> tgo:TGME49_068710 elongation factor Tu GTP-binding domain-containing
protein (EC:2.7.7.4); K03234 elongation factor 2
Length=1697
Score = 101 bits (251), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 47/67 (70%), Positives = 56/67 (83%), Gaps = 0/67 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+ ILAHVDHGKT LSD LL+ NG I+EASA K+R+LDSR+DEQ RQITIKASVVSL
Sbjct 16 IRNVCILAHVDHGKTCLSDRLLSINGLISEASAGKVRYLDSREDEQRRQITIKASVVSLF 75
Query 107 YQRRPQG 113
++R G
Sbjct 76 FRRPDTG 82
> xla:432029 eftud1, MGC83880; elongation factor Tu GTP binding
domain containing 1; K14536 ribosome assembly protein 1 [EC:3.6.5.-]
Length=310
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 40/72 (55%), Positives = 54/72 (75%), Gaps = 0/72 (0%)
Query 46 FVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSL 105
++RNI ILAHVDHGKT+L+DCL++ NG I+ KLR+LDSR+DEQ R IT+K+S +SL
Sbjct 18 YIRNICILAHVDHGKTTLADCLISNNGIISNRLVGKLRYLDSREDEQIRGITMKSSAISL 77
Query 106 LYQRRPQGPLAN 117
Y+ + L N
Sbjct 78 HYKDGEEEYLIN 89
> mmu:101592 Eftud1, 4932434J20Rik, 6030468D11Rik, AI451340, AU019507,
AU022896, D7Ertd791e; elongation factor Tu GTP binding
domain containing 1; K14536 ribosome assembly protein 1
[EC:3.6.5.-]
Length=1127
Score = 87.8 bits (216), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 37/61 (60%), Positives = 51/61 (83%), Gaps = 0/61 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RNI +LAHVDHGKT+L+DCL+++NG I+ A KLR++DSR+DEQ R IT+K+S +SL
Sbjct 19 IRNICVLAHVDHGKTTLADCLISSNGIISSRLAGKLRYMDSREDEQVRGITMKSSAISLH 78
Query 107 Y 107
Y
Sbjct 79 Y 79
> dre:568741 Elongation FacTor family member (eft-2)-like; K14536
ribosome assembly protein 1 [EC:3.6.5.-]
Length=1115
Score = 87.0 bits (214), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 38/61 (62%), Positives = 51/61 (83%), Gaps = 0/61 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+ ILAHVDHGKT+L+DCL+A+NG I+ A KLR+LDSR+DEQ R IT+K+S +SL
Sbjct 19 IRNLCILAHVDHGKTTLADCLVASNGIISSRLAGKLRYLDSREDEQIRGITMKSSAISLH 78
Query 107 Y 107
+
Sbjct 79 F 79
> tpv:TP01_1088 elongation factor Tu; K03234 elongation factor
2
Length=1210
Score = 84.7 bits (208), Expect = 8e-17, Method: Composition-based stats.
Identities = 38/65 (58%), Positives = 51/65 (78%), Gaps = 0/65 (0%)
Query 45 EFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVS 104
E +RN+ LAHVDHGKT+LSD L+++ G I+E + KLR+LD+R DEQ R ITIK+S +S
Sbjct 12 ENIRNVCFLAHVDHGKTTLSDSLISSVGIISEKLSGKLRYLDNRDDEQRRMITIKSSSIS 71
Query 105 LLYQR 109
LLY +
Sbjct 72 LLYSK 76
> sce:YNL163C RIA1, EFL1; Cytoplasmic GTPase involved in biogenesis
of the 60S ribosome; has similarity to translation elongation
factor 2 (Eft1p and Eft2p); K14536 ribosome assembly
protein 1 [EC:3.6.5.-]
Length=1110
Score = 84.3 bits (207), Expect = 8e-17, Method: Composition-based stats.
Identities = 41/74 (55%), Positives = 57/74 (77%), Gaps = 2/74 (2%)
Query 42 SSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKAS 101
+ P +RNI I+AHVDHGKTSLSD LLA+NG I++ A K+RFLD+R DEQ R IT+++S
Sbjct 14 NDPSCIRNICIVAHVDHGKTSLSDSLLASNGIISQRLAGKIRFLDARPDEQLRGITMESS 73
Query 102 VVSLLYQ--RRPQG 113
+SL ++ R+ +G
Sbjct 74 AISLYFRVLRKQEG 87
> hsa:100505780 hypothetical protein LOC100505780
Length=263
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 36/61 (59%), Positives = 50/61 (81%), Gaps = 0/61 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RNI +LAHVD GKT+L+DCL+++NG I+ A KLR++DSR+DEQ R IT+K+S +SL
Sbjct 150 IRNICVLAHVDRGKTTLADCLISSNGIISSHLAGKLRYMDSREDEQIRGITMKSSAISLH 209
Query 107 Y 107
Y
Sbjct 210 Y 210
> hsa:100508498 hypothetical protein LOC100508498
Length=263
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 36/61 (59%), Positives = 50/61 (81%), Gaps = 0/61 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RNI +LAHVD GKT+L+DCL+++NG I+ A KLR++DSR+DEQ R IT+K+S +SL
Sbjct 150 IRNICVLAHVDRGKTTLADCLISSNGIISSHLAGKLRYMDSREDEQIRGITMKSSAISLH 209
Query 107 Y 107
Y
Sbjct 210 Y 210
> cpv:cgd6_3960 elongation factor-like protein
Length=1100
Score = 80.9 bits (198), Expect = 1e-15, Method: Composition-based stats.
Identities = 35/62 (56%), Positives = 50/62 (80%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+ I+AHVDHGKT+L+D LLA+N ++ SA +R+LDSR+DEQ R IT+K+S VSL
Sbjct 5 IRNVCIIAHVDHGKTTLADYLLASNNILSNKSAGTIRYLDSREDEQYRLITMKSSAVSLK 64
Query 107 YQ 108
++
Sbjct 65 FK 66
> pfa:MAL13P1.243 elongation factor Tu, putative
Length=1394
Score = 78.6 bits (192), Expect = 5e-15, Method: Composition-based stats.
Identities = 35/66 (53%), Positives = 51/66 (77%), Gaps = 0/66 (0%)
Query 40 NFSSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIK 99
+ S + +RNI ILAHVDHGKT+L D L+++N I+E + K+++LDSR+DEQ RQIT+K
Sbjct 6 HLSDNDKIRNICILAHVDHGKTTLVDNLISSNKIISEKNIGKIKYLDSREDEQKRQITMK 65
Query 100 ASVVSL 105
+S + L
Sbjct 66 SSSILL 71
> cel:K10C3.5 hypothetical protein; K14536 ribosome assembly protein
1 [EC:3.6.5.-]
Length=894
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 34/62 (54%), Positives = 48/62 (77%), Gaps = 0/62 (0%)
Query 45 EFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVS 104
E +RN+ ++AHVDHGKTS +D L++ N I+ A KLR++DSR+DEQ R IT+K+S +S
Sbjct 18 EHIRNVCLVAHVDHGKTSFADSLVSANAVISSRMAGKLRYMDSREDEQTRGITMKSSGIS 77
Query 105 LL 106
LL
Sbjct 78 LL 79
> xla:380348 eef2.1, MGC53560, eef-2, eef2, ef2; eukaryotic translation
elongation factor 2, gene 1; K03234 elongation factor
2
Length=858
Score = 76.3 bits (186), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/62 (51%), Positives = 47/62 (75%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+S++AHVDHGK++L+D L+ G I A A + RF D+R+DEQ R ITIK++ +SL
Sbjct 19 IRNMSVIAHVDHGKSTLTDSLVCKAGIIASARAGETRFTDTRKDEQERCITIKSTAISLF 78
Query 107 YQ 108
Y+
Sbjct 79 YE 80
> xla:100505433 hypothetical protein LOC100505433; K03234 elongation
factor 2
Length=858
Score = 76.3 bits (186), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/62 (51%), Positives = 47/62 (75%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+S++AHVDHGK++L+D L+ G I A A + RF D+R+DEQ R ITIK++ +SL
Sbjct 19 IRNMSVIAHVDHGKSTLTDSLVCKAGIIASARAGETRFTDTRKDEQERCITIKSTAISLF 78
Query 107 YQ 108
Y+
Sbjct 79 YE 80
> mmu:13629 Eef2, Ef-2, MGC98463; eukaryotic translation elongation
factor 2; K03234 elongation factor 2
Length=858
Score = 76.3 bits (186), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/62 (51%), Positives = 47/62 (75%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+S++AHVDHGK++L+D L+ G I A A + RF D+R+DEQ R ITIK++ +SL
Sbjct 19 IRNMSVIAHVDHGKSTLTDSLVCKAGIIASARAGETRFTDTRKDEQERCITIKSTAISLF 78
Query 107 YQ 108
Y+
Sbjct 79 YE 80
> hsa:1938 EEF2, EEF-2, EF2; eukaryotic translation elongation
factor 2; K03234 elongation factor 2
Length=858
Score = 76.3 bits (186), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/62 (51%), Positives = 47/62 (75%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+S++AHVDHGK++L+D L+ G I A A + RF D+R+DEQ R ITIK++ +SL
Sbjct 19 IRNMSVIAHVDHGKSTLTDSLVCKAGIIASARAGETRFTDTRKDEQERCITIKSTAISLF 78
Query 107 YQ 108
Y+
Sbjct 79 YE 80
> xla:398780 hypothetical protein MGC68699
Length=350
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 32/62 (51%), Positives = 47/62 (75%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+S++AHVDHGK++L+D L+ G I A A + RF D+R+DEQ R ITIK++ +SL
Sbjct 19 IRNMSVIAHVDHGKSTLTDSLVCKAGIIASARAGETRFTDTRKDEQERCITIKSTAISLY 78
Query 107 YQ 108
Y+
Sbjct 79 YE 80
> dre:336168 eef2l2, fj53d02, si:ch211-113n10.4, wu:fj53d02; eukaryotic
translation elongation factor 2, like 2; K03234 elongation
factor 2
Length=861
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/62 (51%), Positives = 47/62 (75%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+S++AHVDHGK++L+D L+ G I A A + RF D+R+DEQ R ITIK++ +SL
Sbjct 19 IRNMSVIAHVDHGKSTLTDSLVCKAGIIASARAGETRFTDTRKDEQERCITIKSTAISLY 78
Query 107 YQ 108
Y+
Sbjct 79 YE 80
> dre:326929 eef2b, EEF2, MGC63584, eef2l, fe49h02, wu:fe49h02,
zgc:63584; eukaryotic translation elongation factor 2b; K03234
elongation factor 2
Length=858
Score = 75.9 bits (185), Expect = 4e-14, Method: Composition-based stats.
Identities = 31/62 (50%), Positives = 48/62 (77%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+S++AHVDHGK++L+D L++ G I A A + RF D+R+DEQ R ITIK++ +S+
Sbjct 19 IRNMSVIAHVDHGKSTLTDSLVSKAGIIASARAGETRFTDTRKDEQERCITIKSTAISMY 78
Query 107 YQ 108
Y+
Sbjct 79 YE 80
> cel:F25H5.4 eft-2; Elongation FacTor family member (eft-2);
K03234 elongation factor 2
Length=852
Score = 75.5 bits (184), Expect = 4e-14, Method: Composition-based stats.
Identities = 35/76 (46%), Positives = 53/76 (69%), Gaps = 6/76 (7%)
Query 33 AQMARKLNFSSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQ 92
A M RK N +RN+S++AHVDHGK++L+D L++ G I + A + RF D+R+DEQ
Sbjct 11 ALMDRKRN------IRNMSVIAHVDHGKSTLTDSLVSKAGIIAGSKAGETRFTDTRKDEQ 64
Query 93 NRQITIKASVVSLLYQ 108
R ITIK++ +SL ++
Sbjct 65 ERCITIKSTAISLFFE 80
> pfa:PF14_0486 elongation factor 2; K03234 elongation factor
2
Length=832
Score = 73.6 bits (179), Expect = 1e-13, Method: Composition-based stats.
Identities = 30/62 (48%), Positives = 48/62 (77%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+S++AHVDHGK++L+D L++ G I+ +A RF D+RQDEQ R ITIK++ +S+
Sbjct 19 IRNMSVIAHVDHGKSTLTDSLVSKAGIISSKNAGDARFTDTRQDEQERCITIKSTGISMY 78
Query 107 YQ 108
++
Sbjct 79 FE 80
> sce:YOR133W EFT1; Eft1p; K03234 elongation factor 2
Length=842
Score = 73.6 bits (179), Expect = 2e-13, Method: Composition-based stats.
Identities = 32/59 (54%), Positives = 46/59 (77%), Gaps = 0/59 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSL 105
VRN+S++AHVDHGK++L+D L+ G I+ A A + RF D+R+DEQ R ITIK++ +SL
Sbjct 19 VRNMSVIAHVDHGKSTLTDSLVQRAGIISAAKAGEARFTDTRKDEQERGITIKSTAISL 77
> sce:YDR385W EFT2; Eft2p; K03234 elongation factor 2
Length=842
Score = 73.6 bits (179), Expect = 2e-13, Method: Composition-based stats.
Identities = 32/59 (54%), Positives = 46/59 (77%), Gaps = 0/59 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSL 105
VRN+S++AHVDHGK++L+D L+ G I+ A A + RF D+R+DEQ R ITIK++ +SL
Sbjct 19 VRNMSVIAHVDHGKSTLTDSLVQRAGIISAAKAGEARFTDTRKDEQERGITIKSTAISL 77
> tgo:TGME49_005470 elongation factor 2, putative ; K03234 elongation
factor 2
Length=832
Score = 72.4 bits (176), Expect = 3e-13, Method: Composition-based stats.
Identities = 30/66 (45%), Positives = 50/66 (75%), Gaps = 0/66 (0%)
Query 43 SPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASV 102
+P+ +RN+S++AHVDHGK++L+D L++ G I+ +A RF D+R DEQ R ITIK++
Sbjct 15 NPKNIRNMSVIAHVDHGKSTLTDSLVSKAGIISAKAAGDARFTDTRADEQERCITIKSTG 74
Query 103 VSLLYQ 108
+S+ ++
Sbjct 75 ISMYFE 80
> cpv:cgd8_2930 Eft2p GTpase; translation elongation factor 2
(EF-2) ; K03234 elongation factor 2
Length=836
Score = 72.4 bits (176), Expect = 4e-13, Method: Composition-based stats.
Identities = 31/65 (47%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 44 PEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVV 103
P +RN+S++AHVDHGK++L+D L+ G I +A RF D+R DEQ R ITIK++ +
Sbjct 20 PHNIRNMSVIAHVDHGKSTLTDSLVCKAGIIASKAAGDARFTDTRADEQERCITIKSTGI 79
Query 104 SLLYQ 108
SL ++
Sbjct 80 SLFFE 84
> tpv:TP01_0529 elongation factor 2; K03234 elongation factor
2
Length=825
Score = 72.4 bits (176), Expect = 4e-13, Method: Composition-based stats.
Identities = 30/66 (45%), Positives = 49/66 (74%), Gaps = 0/66 (0%)
Query 43 SPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASV 102
+P+ +RN+S++AHVDHGK++L+D L++ G I +A RF D+R DEQ R ITIK++
Sbjct 7 NPKNIRNMSVIAHVDHGKSTLTDSLVSKAGIIAAKNAGDARFTDTRADEQERCITIKSTG 66
Query 103 VSLLYQ 108
+S+ ++
Sbjct 67 ISMYFE 72
> bbo:BBOV_I003090 19.m02240; elongation factor 2, EF-2; K03234
elongation factor 2
Length=833
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 30/66 (45%), Positives = 49/66 (74%), Gaps = 0/66 (0%)
Query 43 SPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASV 102
+P+ +RN+S++AHVDHGK++L+D L++ G I +A RF D+R DEQ R ITIK++
Sbjct 15 NPKNIRNMSVIAHVDHGKSTLTDSLVSKAGIIAAKNAGDARFTDTRADEQERCITIKSTG 74
Query 103 VSLLYQ 108
+S+ ++
Sbjct 75 ISMYFE 80
> xla:446712 eef2.2, MGC84492, eef-2, ef2, eft-2; eukaryotic translation
elongation factor 2, gene 2
Length=850
Score = 71.2 bits (173), Expect = 8e-13, Method: Composition-based stats.
Identities = 30/59 (50%), Positives = 45/59 (76%), Gaps = 0/59 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSL 105
+RN+S++AHVDHGK++L+D L+ G I ++ A RF D+R+DEQ R ITIK++ +SL
Sbjct 19 IRNMSVIAHVDHGKSTLTDSLVCKAGIIADSRAGDARFTDTRKDEQERCITIKSTAISL 77
> dre:792182 eef2a.1, MGC113191, si:dkey-110c1.3, zgc:113191;
eukaryotic translation elongation factor 2a, tandem duplicate
1; K03234 elongation factor 2
Length=854
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 31/76 (40%), Positives = 52/76 (68%), Gaps = 6/76 (7%)
Query 33 AQMARKLNFSSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQ 92
A M +K N +RN+S++ DHGK++L+D L++ G ++ A A + RF+D+R+DEQ
Sbjct 11 ATMDKKSN------IRNMSVIGAFDHGKSTLTDWLVSEAGIVSSARAGETRFMDTRRDEQ 64
Query 93 NRQITIKASVVSLLYQ 108
R ITIK++ +S+ Y+
Sbjct 65 ERCITIKSTAISIFYE 80
> dre:568904 eef2a.2, si:dkey-110c1.4; eukaryotic translation
elongation factor 2a, tandem duplicate 2; K03234 elongation
factor 2
Length=853
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 31/76 (40%), Positives = 52/76 (68%), Gaps = 6/76 (7%)
Query 33 AQMARKLNFSSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQ 92
A M +K N +RN+S++ DHGK++L+D L++ G ++ A A + RF+D+R+DEQ
Sbjct 11 ATMDKKSN------IRNMSVIGAFDHGKSTLTDWLVSKAGIVSSACAGETRFMDTRRDEQ 64
Query 93 NRQITIKASVVSLLYQ 108
R ITIK++ +S+ Y+
Sbjct 65 ERCITIKSTAISIFYE 80
> ath:AT1G56070 LOS1; LOS1; copper ion binding / translation elongation
factor/ translation factor, nucleic acid binding;
K03234 elongation factor 2
Length=843
Score = 69.7 bits (169), Expect = 3e-12, Method: Composition-based stats.
Identities = 30/62 (48%), Positives = 45/62 (72%), Gaps = 0/62 (0%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN+S++AHVDHGK++L+D L+A G I + A +R D+R DE R ITIK++ +SL
Sbjct 19 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKSTGISLY 78
Query 107 YQ 108
Y+
Sbjct 79 YE 80
> bbo:BBOV_IV003290 21.m02927; Elongation factor Tu-like protein;
K03234 elongation factor 2
Length=1222
Score = 68.9 bits (167), Expect = 4e-12, Method: Composition-based stats.
Identities = 40/74 (54%), Positives = 56/74 (75%), Gaps = 1/74 (1%)
Query 35 MARKLNF-SSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQN 93
MA++ + S E +RN+ LAHVDHGKT+LSD L+++ G I+E + +LR+LD+R DEQ
Sbjct 1 MAQQTDLLKSTEHIRNVCFLAHVDHGKTTLSDSLISSIGIISERMSGRLRYLDNRDDEQR 60
Query 94 RQITIKASVVSLLY 107
R ITIK+S +SLLY
Sbjct 61 RMITIKSSSISLLY 74
> tgo:TGME49_107980 GTP-binding protein lepA, putative (EC:2.7.7.4);
K03596 GTP-binding protein LepA
Length=638
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/66 (54%), Positives = 46/66 (69%), Gaps = 3/66 (4%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
VRN SI+AH+DHGK++LSD LL G I++ AK +FLDS + E R IT+KA SLL
Sbjct 34 VRNFSIIAHIDHGKSTLSDRLLELTGTISQ--GAKAQFLDSLEVETTRGITVKAQTCSLL 91
Query 107 YQRRPQ 112
Y R P+
Sbjct 92 Y-RHPK 96
> ath:AT3G22980 elongation factor Tu family protein; K14536 ribosome
assembly protein 1 [EC:3.6.5.-]
Length=1015
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 35/64 (54%), Positives = 47/64 (73%), Gaps = 2/64 (3%)
Query 47 VRNISILAHVDHGKTSLSDCLLATN--GFITEASAAKLRFLDSRQDEQNRQITIKASVVS 104
VRNI ILAHVDHGKT+L+D L+A++ G + A KLRF+D +EQ R IT+K+S +S
Sbjct 9 VRNICILAHVDHGKTTLADHLIASSGGGVLHPRLAGKLRFMDYLDEEQRRAITMKSSSIS 68
Query 105 LLYQ 108
L Y+
Sbjct 69 LKYK 72
> ath:AT3G12915 GTP binding / GTPase; K03234 elongation factor
2
Length=820
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 28/59 (47%), Positives = 43/59 (72%), Gaps = 0/59 (0%)
Query 50 ISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLLYQ 108
+S++AHVDHGK++L+D L+A G I + +A +R D+R DE R ITIK++ +SL Y+
Sbjct 1 MSVIAHVDHGKSTLTDSLVAAAGIIAQETAGDVRMTDTRADEAERGITIKSTGISLYYE 59
> dre:553030 guf1, im:6906935, wu:fc88c11; GUF1 GTPase homolog
(S. cerevisiae)
Length=672
Score = 64.3 bits (155), Expect = 1e-10, Method: Composition-based stats.
Identities = 36/91 (39%), Positives = 54/91 (59%), Gaps = 1/91 (1%)
Query 27 VAASAAAQMARKLNFSSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLD 86
+++SA + ++ E +RN SI+AH+DHGK++L+D LL G I + ++ K + LD
Sbjct 54 LSSSATEKETFDMSKFPAERIRNFSIIAHIDHGKSTLADRLLEMTGAIAKTASNK-QVLD 112
Query 87 SRQDEQNRQITIKASVVSLLYQRRPQGPLAN 117
Q E+ R IT+KA SL YQ Q L N
Sbjct 113 KLQVERERGITVKAQTASLFYQHHGQTYLLN 143
> pfa:PF07_0062 GTP-binding translation elongation factor tu family
protein, putative
Length=754
Score = 61.2 bits (147), Expect = 9e-10, Method: Composition-based stats.
Identities = 32/72 (44%), Positives = 43/72 (59%), Gaps = 4/72 (5%)
Query 36 ARKLNFSSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQ 95
++KLN PE +RN++I+AHVDHGKT+L D LL G IT R +D E+ R
Sbjct 87 SKKLNIIDPEKIRNVAIIAHVDHGKTTLVDKLLRQGGEITNND----RIMDHNDLEKERG 142
Query 96 ITIKASVVSLLY 107
ITI + V + Y
Sbjct 143 ITIMSKVTRIKY 154
> mmu:231279 Guf1, 4631409J12, AA407526, EF-4; GUF1 GTPase homolog
(S. cerevisiae)
Length=651
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 35/79 (44%), Positives = 48/79 (60%), Gaps = 3/79 (3%)
Query 31 AAAQMARKLNFSS--PEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSR 88
+AA++ K + S E +RN SI+AHVDHGK++L+D LL G I + K + LD
Sbjct 32 SAAELKEKPDMSRFPVEDIRNFSIIAHVDHGKSTLADRLLELTGTIDKTKKNK-QVLDKL 90
Query 89 QDEQNRQITIKASVVSLLY 107
Q E+ R IT+KA SL Y
Sbjct 91 QVERERGITVKAQTASLFY 109
> hsa:60558 GUF1, EF-4, FLJ13220; GUF1 GTPase homolog (S. cerevisiae)
Length=669
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 35/79 (44%), Positives = 48/79 (60%), Gaps = 3/79 (3%)
Query 31 AAAQMARKLNFSS--PEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSR 88
++A+ KL+ S E +RN SI+AHVDHGK++L+D LL G I + K + LD
Sbjct 50 SSAEFKEKLDMSRFPVENIRNFSIVAHVDHGKSTLADRLLELTGTIDKTKNNK-QVLDKL 108
Query 89 QDEQNRQITIKASVVSLLY 107
Q E+ R IT+KA SL Y
Sbjct 109 QVERERGITVKAQTASLFY 127
> ath:AT5G39900 GTP binding / GTPase/ translation elongation factor
Length=663
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 43/66 (65%), Gaps = 1/66 (1%)
Query 45 EFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVS 104
E +RN SI+AH+DHGK++L+D L+ G I + + ++LD Q E+ R IT+KA +
Sbjct 64 EKIRNFSIIAHIDHGKSTLADRLMELTGTIKKGH-GQPQYLDKLQVERERGITVKAQTAT 122
Query 105 LLYQRR 110
+ Y+ +
Sbjct 123 MFYENK 128
> cel:ZK1236.1 hypothetical protein
Length=645
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 38/103 (36%), Positives = 58/103 (56%), Gaps = 10/103 (9%)
Query 17 FPGELKLTGCVAASAAAQMARKLNFS--SPEFVRNISILAHVDHGKTSLSDCLLATNGFI 74
F G+L+++ C S A + +N S +P+ +RN I+AHVDHGK++L+D LL G +
Sbjct 11 FIGKLRVS-CKHYSTAGDPTKLVNLSEFTPDKIRNFGIVAHVDHGKSTLADRLLEMCGAV 69
Query 75 TEASAAKLRFLDSRQDEQNRQITIKASVVSLLYQRRPQGPLAN 117
+ + LD Q E+ R IT+KA +L R +G L N
Sbjct 70 ---PPGQKQMLDKLQVERERGITVKAQTAAL----RHRGYLLN 105
> sce:YLR289W GUF1; Guf1p
Length=645
Score = 55.1 bits (131), Expect = 7e-08, Method: Composition-based stats.
Identities = 31/69 (44%), Positives = 42/69 (60%), Gaps = 1/69 (1%)
Query 45 EFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVS 104
E RN SI+AHVDHGK++LSD LL I + +A + LD + E+ R ITIKA S
Sbjct 44 ENYRNFSIVAHVDHGKSTLSDRLLEITHVI-DPNARNKQVLDKLEVERERGITIKAQTCS 102
Query 105 LLYQRRPQG 113
+ Y+ + G
Sbjct 103 MFYKDKRTG 111
> ath:AT5G13650 elongation factor family protein
Length=676
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 43/80 (53%), Gaps = 0/80 (0%)
Query 29 ASAAAQMARKLNFSSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSR 88
A A+ +K + VRNI+I+AHVDHGKT+L D +L + + R +DS
Sbjct 65 AEPASVEVKKKQLDRRDNVRNIAIVAHVDHGKTTLVDSMLRQAKVFRDNQVMQERIMDSN 124
Query 89 QDEQNRQITIKASVVSLLYQ 108
E+ R ITI + S+ Y+
Sbjct 125 DLERERGITILSKNTSITYK 144
> ath:AT5G25230 elongation factor Tu family protein; K12852 116
kDa U5 small nuclear ribonucleoprotein component
Length=973
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 43/70 (61%), Gaps = 3/70 (4%)
Query 42 SSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAK---LRFLDSRQDEQNRQITI 98
S+P VRN++++ H+ HGKT D L+ ++ +A +R+ D+R DEQ R I+I
Sbjct 119 SNPALVRNVALVGHLQHGKTVFMDMLVEQTHRMSTFNAENDKHMRYTDTRVDEQERNISI 178
Query 99 KASVVSLLYQ 108
KA +SL+ +
Sbjct 179 KAVPMSLVLE 188
> ath:AT1G06220 MEE5; MEE5 (MATERNAL EFFECT EMBRYO ARREST 5);
GTP binding / GTPase/ translation elongation factor/ translation
factor, nucleic acid binding; K12852 116 kDa U5 small nuclear
ribonucleoprotein component
Length=987
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 25/70 (35%), Positives = 43/70 (61%), Gaps = 3/70 (4%)
Query 42 SSPEFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAK---LRFLDSRQDEQNRQITI 98
S+P VRN++++ H+ HGKT D L+ ++ +A +++ D+R DEQ R I+I
Sbjct 133 SNPALVRNVALVGHLQHGKTVFMDMLVEQTHHMSTFNAKNEKHMKYTDTRVDEQERNISI 192
Query 99 KASVVSLLYQ 108
KA +SL+ +
Sbjct 193 KAVPMSLVLE 202
> bbo:BBOV_II004050 18.m06335; u5 small nuclear ribonuclear protein;
K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=999
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 28/79 (35%), Positives = 43/79 (54%), Gaps = 13/79 (16%)
Query 44 PEFVRNISILAHVDHGKTSLSDCLL---------ATNGFITEASAAKLRFLDSRQDEQNR 94
P+F+RN+ I HGKT+L D + GF T + R+ D+R DEQ R
Sbjct 130 PQFIRNVCICGDFHHGKTTLIDRFINYSRYPAPDCAEGFDTSFT----RYTDTRLDEQAR 185
Query 95 QITIKASVVSLLYQRRPQG 113
Q++IK++ +SL++Q G
Sbjct 186 QMSIKSTPISLVFQTETGG 204
> eco:b3871 typA, bipA, ECK3864, JW5571, yihK; GTP-binding protein;
K06207 GTP-binding protein
Length=607
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 27/56 (48%), Positives = 36/56 (64%), Gaps = 0/56 (0%)
Query 45 EFVRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKA 100
E +RNI+I+AHVDHGKT+L D LL +G + + R +DS E+ R ITI A
Sbjct 3 EKLRNIAIIAHVDHGKTTLVDKLLQQSGTFDSRAETQERVMDSNDLEKERGITILA 58
> tpv:TP04_0166 U5 small nuclear ribonucleoprotein; K12852 116
kDa U5 small nuclear ribonucleoprotein component
Length=1028
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 27/70 (38%), Positives = 41/70 (58%), Gaps = 5/70 (7%)
Query 44 PEFVRNISILAHVDHGKTSLSDCLL-----ATNGFITEASAAKLRFLDSRQDEQNRQITI 98
PEF+RNI I GKT+L D L+ + T + R+ DSR DEQ R+++I
Sbjct 136 PEFIRNICICGGFHDGKTTLIDRLIEFSRYQSTSLDTRKNPEFTRYTDSRLDEQARELSI 195
Query 99 KASVVSLLYQ 108
K++ +SL++Q
Sbjct 196 KSTPISLIFQ 205
> ath:AT5G08650 GTP-binding protein LepA, putative; K03596 GTP-binding
protein LepA
Length=681
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 28/67 (41%), Positives = 43/67 (64%), Gaps = 3/67 (4%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIK--ASVVS 104
+RN SI+AH+DHGK++L+D LL G + + K +FLD+ E+ R ITIK A+ +
Sbjct 86 IRNFSIIAHIDHGKSTLADKLLQVTGTV-QNRDMKEQFLDNMDLERERGITIKLQAARMR 144
Query 105 LLYQRRP 111
+Y+ P
Sbjct 145 YVYEDTP 151
> eco:b2569 lepA, ECK2567, JW2553; back-translocating elongation
Factor EF4, GTPase; K03596 GTP-binding protein LepA
Length=599
Score = 52.0 bits (123), Expect = 6e-07, Method: Composition-based stats.
Identities = 27/62 (43%), Positives = 42/62 (67%), Gaps = 1/62 (1%)
Query 47 VRNISILAHVDHGKTSLSDCLLATNGFITEASAAKLRFLDSRQDEQNRQITIKASVVSLL 106
+RN SI+AH+DHGK++LSD ++ G +++ + + LDS E+ R ITIKA V+L
Sbjct 4 IRNFSIIAHIDHGKSTLSDRIIQICGGLSDRE-MEAQVLDSMDLERERGITIKAQSVTLD 62
Query 107 YQ 108
Y+
Sbjct 63 YK 64
> cpv:cgd3_3880 Snu114p GTpase, U5 snRNP-specific protein, 116
kDa ; K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=1035
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 28/65 (43%), Positives = 41/65 (63%), Gaps = 3/65 (4%)
Query 45 EFVRNISILAHVDHGKTSLSDCLLA-TNGFITEASAAKL--RFLDSRQDEQNRQITIKAS 101
EFVRNI + + GKT+ D L+ T+ + + L R+ DSR+DEQ+R I+IKAS
Sbjct 173 EFVRNICFIGEIHSGKTTFLDMLIKNTHSYKGDKKNIPLPERYCDSRKDEQDRGISIKAS 232
Query 102 VVSLL 106
+SL+
Sbjct 233 PISLV 237
Lambda K H
0.316 0.129 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2296762580
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40