bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2931_orf2
Length=202
Score E
Sequences producing significant alignments: (Bits) Value
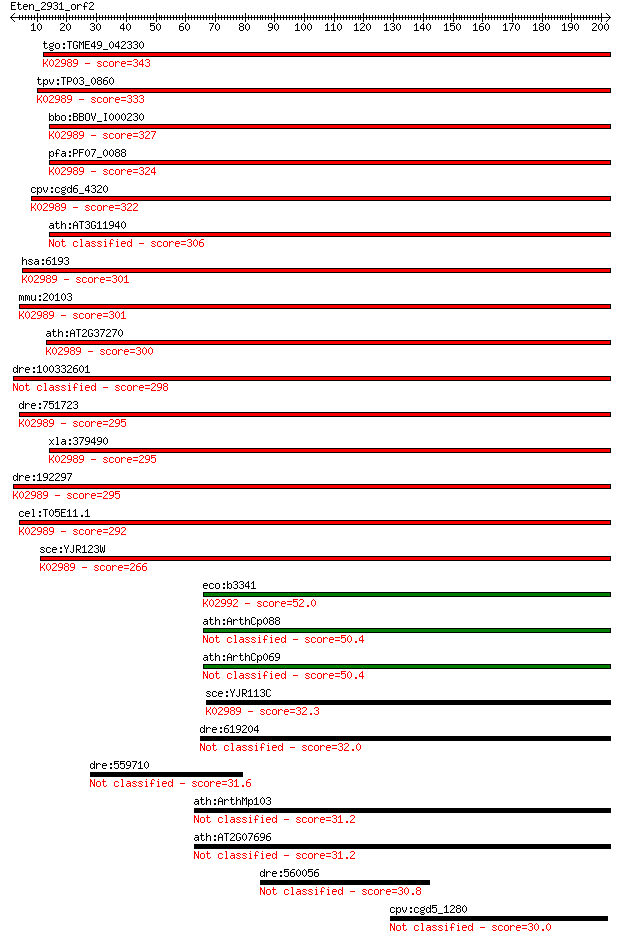
tgo:TGME49_042330 40S ribosomal protein S5, putative ; K02989 ... 343 3e-94
tpv:TP03_0860 40S ribosomal protein S5; K02989 small subunit r... 333 2e-91
bbo:BBOV_I000230 16.m00772; 40S ribosomal protein S5; K02989 s... 327 1e-89
pfa:PF07_0088 40S ribosomal protein S5, putative; K02989 small... 324 2e-88
cpv:cgd6_4320 40S ribosomal protein S5 ; K02989 small subunit ... 322 5e-88
ath:AT3G11940 ATRPS5A (RIBOSOMAL PROTEIN 5A); structural const... 306 3e-83
hsa:6193 RPS5; ribosomal protein S5; K02989 small subunit ribo... 301 9e-82
mmu:20103 Rps5, AA617411; ribosomal protein S5; K02989 small s... 301 1e-81
ath:AT2G37270 ATRPS5B (RIBOSOMAL PROTEIN 5B); structural const... 300 2e-81
dre:100332601 ribosomal protein S5-like 298 1e-80
dre:751723 MGC153322; zgc:153322; K02989 small subunit ribosom... 295 5e-80
xla:379490 rps5, MGC64490; ribosomal protein S5; K02989 small ... 295 5e-80
dre:192297 rps5, chunp6933, fc74e05, fj57h03, wu:fc74e05, wu:f... 295 7e-80
cel:T05E11.1 rps-5; Ribosomal Protein, Small subunit family me... 292 4e-79
sce:YJR123W RPS5; Rps5p; K02989 small subunit ribosomal protei... 266 3e-71
eco:b3341 rpsG, ECK3328, JW3303; 30S ribosomal subunit protein... 52.0 1e-06
ath:ArthCp088 rps7; 30S ribosomal protein S7 50.4 4e-06
ath:ArthCp069 rps7; 30S ribosomal protein S7 50.4 4e-06
sce:YJR113C RSM7; Mitochondrial ribosomal protein of the small... 32.3 1.0
dre:619204 mrps7, MGC110673, zgc:110673; mitochondrial ribosom... 32.0 1.6
dre:559710 tapt1; similar to FLJ90013 protein 31.6 1.9
ath:ArthMp103 rps7; ribosomal protein S7 31.2 2.8
ath:AT2G07696 ribosomal protein S7 family protein 31.2 2.8
dre:560056 znf704, si:ch211-203l9., si:ch211-203l9.2, si:ch211... 30.8 3.5
cpv:cgd5_1280 tfc4p like TFIIIC subunit; TPR repeat containing... 30.0 6.1
> tgo:TGME49_042330 40S ribosomal protein S5, putative ; K02989
small subunit ribosomal protein S5e
Length=192
Score = 343 bits (880), Expect = 3e-94, Method: Compositional matrix adjust.
Identities = 160/191 (83%), Positives = 176/191 (92%), Gaps = 0/191 (0%)
Query 12 ATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLVERLV 71
AT+ KLFGRW+YDDV VSDLSLVDY+AVK KA V PHTAGRYQKKRFRKA CP+VERLV
Sbjct 2 ATEAKLFGRWSYDDVNVSDLSLVDYIAVKDKACVFVPHTAGRYQKKRFRKAMCPIVERLV 61
Query 72 NSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAGV 131
NS+MMHGRN+GKK ++RIV+HAF+II LM DKNP+QV V AVE GGPREDSTRIGSAGV
Sbjct 62 NSMMMHGRNNGKKTLSVRIVRHAFEIIHLMTDKNPIQVFVNAVENGGPREDSTRIGSAGV 121
Query 132 VRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKKK 191
VRRQAVDVSPLRRVNQAIYLICTGAR AAFRN+K+++ECLADEIMNCAKESSN+YAIKKK
Sbjct 122 VRRQAVDVSPLRRVNQAIYLICTGARLAAFRNIKTIAECLADEIMNCAKESSNAYAIKKK 181
Query 192 DEIERVAKANR 202
DEIERVAKANR
Sbjct 182 DEIERVAKANR 192
> tpv:TP03_0860 40S ribosomal protein S5; K02989 small subunit
ribosomal protein S5e
Length=193
Score = 333 bits (855), Expect = 2e-91, Method: Compositional matrix adjust.
Identities = 154/193 (79%), Positives = 176/193 (91%), Gaps = 0/193 (0%)
Query 10 MAATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLVER 69
M T+ LF +WTYDDV+++DLSLVD +A++GKA V TPHTAGRYQKKRFRK QCP+VER
Sbjct 1 MTNTEVALFRKWTYDDVSLTDLSLVDCIAIQGKARVFTPHTAGRYQKKRFRKTQCPIVER 60
Query 70 LVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSA 129
LVNS+MMHGRN+GKKL+AIRIV HAF+II +M DKNP+QV V AV+ GGPREDSTRIGSA
Sbjct 61 LVNSMMMHGRNNGKKLKAIRIVNHAFEIIHVMTDKNPVQVFVDAVKNGGPREDSTRIGSA 120
Query 130 GVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIK 189
GVVRRQAVDVSPLRRVNQAIYLICTGAR AAFRN+K+++ECLADEI+NCAKES +SYAIK
Sbjct 121 GVVRRQAVDVSPLRRVNQAIYLICTGARNAAFRNIKTIAECLADEIINCAKESPSSYAIK 180
Query 190 KKDEIERVAKANR 202
KKDEIERVAKANR
Sbjct 181 KKDEIERVAKANR 193
> bbo:BBOV_I000230 16.m00772; 40S ribosomal protein S5; K02989
small subunit ribosomal protein S5e
Length=192
Score = 327 bits (839), Expect = 1e-89, Method: Compositional matrix adjust.
Identities = 153/189 (80%), Positives = 171/189 (90%), Gaps = 0/189 (0%)
Query 14 DFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLVERLVNS 73
+ LF +W YD V++SDLSLVD +A++GKA V TPHTAGRYQKKRFRK CP+VERLVNS
Sbjct 4 ELALFRKWPYDSVSLSDLSLVDCIAIQGKARVFTPHTAGRYQKKRFRKTLCPIVERLVNS 63
Query 74 LMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAGVVR 133
LMMHGRNSGKKL+AIRIV HAF+II LM D+NP+QV V AV+ GGPREDSTRIGSAGVVR
Sbjct 64 LMMHGRNSGKKLKAIRIVMHAFEIIHLMTDQNPIQVFVDAVKNGGPREDSTRIGSAGVVR 123
Query 134 RQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKKKDE 193
RQAVDVSPLRRVNQAIYLICTGAR AAFRN+K+++ECLADEIMNCAKES +SYAIKKKDE
Sbjct 124 RQAVDVSPLRRVNQAIYLICTGARNAAFRNIKTIAECLADEIMNCAKESPSSYAIKKKDE 183
Query 194 IERVAKANR 202
IERVAKANR
Sbjct 184 IERVAKANR 192
> pfa:PF07_0088 40S ribosomal protein S5, putative; K02989 small
subunit ribosomal protein S5e
Length=195
Score = 324 bits (830), Expect = 2e-88, Method: Compositional matrix adjust.
Identities = 150/189 (79%), Positives = 172/189 (91%), Gaps = 0/189 (0%)
Query 14 DFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLVERLVNS 73
D KLF +W+Y+++ ++DLSLVD +AV KA V TPHTAGRYQKKRFRKA CP+VERLVNS
Sbjct 7 DIKLFKKWSYEEINIADLSLVDCIAVSQKACVYTPHTAGRYQKKRFRKALCPIVERLVNS 66
Query 74 LMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAGVVR 133
+MMHGRN+GKKL+AIRIV +AF+II LM +NPLQV V AV++GGPREDSTRIGSAGVVR
Sbjct 67 MMMHGRNNGKKLKAIRIVAYAFEIIHLMTGENPLQVFVNAVQKGGPREDSTRIGSAGVVR 126
Query 134 RQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKKKDE 193
RQAVDVSPLRRVNQAIYLICTGAR AAFRN+KS+SECLA+EI+NCA ESS+SYAIKKKDE
Sbjct 127 RQAVDVSPLRRVNQAIYLICTGARNAAFRNIKSISECLAEEIINCANESSSSYAIKKKDE 186
Query 194 IERVAKANR 202
IERVAKANR
Sbjct 187 IERVAKANR 195
> cpv:cgd6_4320 40S ribosomal protein S5 ; K02989 small subunit
ribosomal protein S5e
Length=207
Score = 322 bits (825), Expect = 5e-88, Method: Compositional matrix adjust.
Identities = 153/195 (78%), Positives = 172/195 (88%), Gaps = 0/195 (0%)
Query 8 ATMAATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLV 67
AT + +D KLFG+W+YDDV VSDLSLVDY+AVK KA V P TAGRYQ KRFRKA CP+V
Sbjct 13 ATFSTSDVKLFGKWSYDDVNVSDLSLVDYIAVKDKACVYAPVTAGRYQVKRFRKALCPIV 72
Query 68 ERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIG 127
ERLV SLMMHGRN+GKK QAIRIV+ AFDII L+ D+NP+QV V AV+ GGPREDST IG
Sbjct 73 ERLVCSLMMHGRNNGKKNQAIRIVRAAFDIIHLVTDQNPIQVFVDAVKNGGPREDSTSIG 132
Query 128 SAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYA 187
SAG VRRQAVDVSPLRRVNQAIYLICTGART AFRN+K+++ECLADEIMNCAKE+ NSYA
Sbjct 133 SAGAVRRQAVDVSPLRRVNQAIYLICTGARTQAFRNIKTIAECLADEIMNCAKENPNSYA 192
Query 188 IKKKDEIERVAKANR 202
IK+KDE+ERVAKANR
Sbjct 193 IKRKDEVERVAKANR 207
> ath:AT3G11940 ATRPS5A (RIBOSOMAL PROTEIN 5A); structural constituent
of ribosome
Length=207
Score = 306 bits (784), Expect = 3e-83, Method: Compositional matrix adjust.
Identities = 141/190 (74%), Positives = 166/190 (87%), Gaps = 1/190 (0%)
Query 14 DFKLFGRWTYDDVTVSDLSLVDYLAVKG-KAAVLTPHTAGRYQKKRFRKAQCPLVERLVN 72
+ KLF RWTYDDVTV+D+SLVDY+ V+ K A PHTAGRY KRFRKAQCP+VERL N
Sbjct 18 EVKLFNRWTYDDVTVTDISLVDYIGVQAAKHATFVPHTAGRYSVKRFRKAQCPIVERLTN 77
Query 73 SLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAGVV 132
SLMMHGRN+GKKL A+RIV+HA +II L++D NP+QV++ A+ GPRED+TRIGSAGVV
Sbjct 78 SLMMHGRNNGKKLMAVRIVKHAMEIIHLLSDLNPIQVIIDAIVNSGPREDATRIGSAGVV 137
Query 133 RRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKKKD 192
RRQAVD+SPLRRVNQAI+LI TGAR AAFRN+K+++ECLADE++N AK SSNSYAIKKKD
Sbjct 138 RRQAVDISPLRRVNQAIFLITTGAREAAFRNIKTIAECLADELINAAKGSSNSYAIKKKD 197
Query 193 EIERVAKANR 202
EIERVAKANR
Sbjct 198 EIERVAKANR 207
> hsa:6193 RPS5; ribosomal protein S5; K02989 small subunit ribosomal
protein S5e
Length=204
Score = 301 bits (771), Expect = 9e-82, Method: Compositional matrix adjust.
Identities = 140/198 (70%), Positives = 166/198 (83%), Gaps = 0/198 (0%)
Query 5 AAAATMAATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQC 64
AA A D KLFG+W+ DDV ++D+SL DY+AVK K A PH+AGRY KRFRKAQC
Sbjct 7 AAPAVAETPDIKLFGKWSTDDVQINDISLQDYIAVKEKYAKYLPHSAGRYAAKRFRKAQC 66
Query 65 PLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDST 124
P+VERL NS+MMHGRN+GKKL +RIV+HAF+II L+ +NPLQVLV A+ GPREDST
Sbjct 67 PIVERLTNSMMMHGRNNGKKLMTVRIVKHAFEIIHLLTGENPLQVLVNAIINSGPREDST 126
Query 125 RIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSN 184
RIG AG VRRQAVDVSPLRRVNQAI+L+CTGAR AAFRN+K+++ECLADE++N AK SSN
Sbjct 127 RIGRAGTVRRQAVDVSPLRRVNQAIWLLCTGAREAAFRNIKTIAECLADELINAAKGSSN 186
Query 185 SYAIKKKDEIERVAKANR 202
SYAIKKKDE+ERVAK+NR
Sbjct 187 SYAIKKKDELERVAKSNR 204
> mmu:20103 Rps5, AA617411; ribosomal protein S5; K02989 small
subunit ribosomal protein S5e
Length=204
Score = 301 bits (771), Expect = 1e-81, Method: Compositional matrix adjust.
Identities = 140/199 (70%), Positives = 166/199 (83%), Gaps = 0/199 (0%)
Query 4 AAAAATMAATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQ 63
AA A D KLFG+W+ DDV ++D+SL DY+AVK K A PH+AGRY KRFRKAQ
Sbjct 6 AATPAVAETPDIKLFGKWSTDDVQINDISLQDYIAVKEKYAKYLPHSAGRYAAKRFRKAQ 65
Query 64 CPLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDS 123
CP+VERL NS+MMHGRN+GKKL +RIV+HAF+II L+ +NPLQVLV A+ GPREDS
Sbjct 66 CPIVERLTNSMMMHGRNNGKKLMTVRIVKHAFEIIHLLTGENPLQVLVNAIINSGPREDS 125
Query 124 TRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESS 183
TRIG AG VRRQAVDVSPLRRVNQAI+L+CTGAR AAFRN+K+++ECLADE++N AK SS
Sbjct 126 TRIGRAGTVRRQAVDVSPLRRVNQAIWLLCTGAREAAFRNIKTIAECLADELINAAKGSS 185
Query 184 NSYAIKKKDEIERVAKANR 202
NSYAIKKKDE+ERVAK+NR
Sbjct 186 NSYAIKKKDELERVAKSNR 204
> ath:AT2G37270 ATRPS5B (RIBOSOMAL PROTEIN 5B); structural constituent
of ribosome; K02989 small subunit ribosomal protein
S5e
Length=207
Score = 300 bits (768), Expect = 2e-81, Method: Compositional matrix adjust.
Identities = 137/191 (71%), Positives = 166/191 (86%), Gaps = 1/191 (0%)
Query 13 TDFKLFGRWTYDDVTVSDLSLVDYLAVK-GKAAVLTPHTAGRYQKKRFRKAQCPLVERLV 71
+ KLF RW++DDV+V+D+SLVDY+ V+ K A PHTAGRY KRFRKAQCP+VERL
Sbjct 17 NEVKLFNRWSFDDVSVTDISLVDYIGVQPSKHATFVPHTAGRYSVKRFRKAQCPIVERLT 76
Query 72 NSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAGV 131
NSLMMHGRN+GKKL A+RIV+HA +II L++D NP+QV++ A+ GPRED+TRIGSAGV
Sbjct 77 NSLMMHGRNNGKKLMAVRIVKHAMEIIHLLSDLNPIQVIIDAIVNSGPREDATRIGSAGV 136
Query 132 VRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKKK 191
VRRQAVD+SPLRRVNQAI+L+ TGAR AAFRN+K+++ECLADE++N AK SSNSYAIKKK
Sbjct 137 VRRQAVDISPLRRVNQAIFLLTTGAREAAFRNIKTIAECLADELINAAKGSSNSYAIKKK 196
Query 192 DEIERVAKANR 202
DEIERVAKANR
Sbjct 197 DEIERVAKANR 207
> dre:100332601 ribosomal protein S5-like
Length=204
Score = 298 bits (762), Expect = 1e-80, Method: Compositional matrix adjust.
Identities = 137/201 (68%), Positives = 167/201 (83%), Gaps = 0/201 (0%)
Query 2 DAAAAAATMAATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRK 61
D AA A + KLFG+W+ DDV ++D+SL DY+AVK K A PH++GRY KRFRK
Sbjct 4 DWEAAPAVAETPEIKLFGKWSTDDVQINDISLQDYIAVKEKYAKYLPHSSGRYAAKRFRK 63
Query 62 AQCPLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPRE 121
AQCP+VER+ NS+MMHGRN+GKKL +RIV+HAF+II L+ +NPLQ+LV A+ GPRE
Sbjct 64 AQCPIVERVTNSMMMHGRNNGKKLLTVRIVKHAFEIIHLLTGENPLQILVNAIINSGPRE 123
Query 122 DSTRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKE 181
DSTRIG AG VRRQAVDVSPLRRVNQAI+L+CTGAR AAFRN+K+++ECLADE++N AK
Sbjct 124 DSTRIGRAGTVRRQAVDVSPLRRVNQAIWLLCTGAREAAFRNIKTIAECLADELINAAKG 183
Query 182 SSNSYAIKKKDEIERVAKANR 202
SSNSYAIKKKDE+ERVAK+NR
Sbjct 184 SSNSYAIKKKDELERVAKSNR 204
> dre:751723 MGC153322; zgc:153322; K02989 small subunit ribosomal
protein S5e
Length=201
Score = 295 bits (756), Expect = 5e-80, Method: Compositional matrix adjust.
Identities = 139/199 (69%), Positives = 165/199 (82%), Gaps = 0/199 (0%)
Query 4 AAAAATMAATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQ 63
A A A + KLFGRW+ +DV VSD+SL DY+AVK A PH+AGRY KRFRKAQ
Sbjct 3 ATAPAPAELPEVKLFGRWSCEDVQVSDMSLQDYIAVKENYAKFIPHSAGRYAAKRFRKAQ 62
Query 64 CPLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDS 123
CP+VER+ NSLMMHGRN+GKKL A+RI++HA +II L+ +NPLQVLV A+ GPREDS
Sbjct 63 CPIVERVTNSLMMHGRNNGKKLMAVRIMKHAMEIIHLLTGENPLQVLVNAIINSGPREDS 122
Query 124 TRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESS 183
TRIG AG VRRQAVDVSPLRRVNQAI+L+CTGAR AAFRN+K+++ECLADE++N AK SS
Sbjct 123 TRIGRAGTVRRQAVDVSPLRRVNQAIWLLCTGAREAAFRNIKTIAECLADELINAAKGSS 182
Query 184 NSYAIKKKDEIERVAKANR 202
NSYAIKKKDE+ERVAK+NR
Sbjct 183 NSYAIKKKDELERVAKSNR 201
> xla:379490 rps5, MGC64490; ribosomal protein S5; K02989 small
subunit ribosomal protein S5e
Length=203
Score = 295 bits (756), Expect = 5e-80, Method: Compositional matrix adjust.
Identities = 135/189 (71%), Positives = 162/189 (85%), Gaps = 0/189 (0%)
Query 14 DFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLVERLVNS 73
+ KLFG+W+ DDV ++D+SL DY+AVK K A PH+ GRY KRFRKAQCP+VERL NS
Sbjct 15 EIKLFGKWSTDDVQINDISLQDYIAVKEKYAKFLPHSGGRYAAKRFRKAQCPIVERLTNS 74
Query 74 LMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAGVVR 133
LMMHGRN+GKKL +RIV+HAF+II L+ +NPLQVLV A+ GPREDSTRIG AG VR
Sbjct 75 LMMHGRNNGKKLMTVRIVKHAFEIIHLLTGENPLQVLVNAIINSGPREDSTRIGRAGTVR 134
Query 134 RQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKKKDE 193
RQAVDVSPLRRVNQAI+L+CTGAR AAFRN+K+++EC+ADE++N AK SSNSYAIKKKDE
Sbjct 135 RQAVDVSPLRRVNQAIWLLCTGAREAAFRNIKTIAECVADELINAAKGSSNSYAIKKKDE 194
Query 194 IERVAKANR 202
+ERVAK+NR
Sbjct 195 LERVAKSNR 203
> dre:192297 rps5, chunp6933, fc74e05, fj57h03, wu:fc74e05, wu:fj57h03;
ribosomal protein S5; K02989 small subunit ribosomal
protein S5e
Length=204
Score = 295 bits (755), Expect = 7e-80, Method: Compositional matrix adjust.
Identities = 136/201 (67%), Positives = 166/201 (82%), Gaps = 0/201 (0%)
Query 2 DAAAAAATMAATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRK 61
D AA A + KLFG+W+ DDV ++D+SL DY+AVK K A PH++GRY KRFRK
Sbjct 4 DWEAAPAVAETPEIKLFGKWSTDDVQINDISLQDYIAVKEKYAKYLPHSSGRYAAKRFRK 63
Query 62 AQCPLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPRE 121
AQCP+VER+ NS+MMHGRN+GKKL +RIV+HAF+II L+ +NPLQ+LV A+ GPRE
Sbjct 64 AQCPIVERVTNSMMMHGRNNGKKLLTVRIVKHAFEIIHLLTGENPLQILVNAIINSGPRE 123
Query 122 DSTRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKE 181
DSTRIG AG VRRQAVDVSPLRRVNQAI+L+CTGAR AAFRN+K+++ECLADE++N AK
Sbjct 124 DSTRIGRAGTVRRQAVDVSPLRRVNQAIWLLCTGAREAAFRNIKTIAECLADELINAAKG 183
Query 182 SSNSYAIKKKDEIERVAKANR 202
S NSYAIKKKDE+ERVAK+NR
Sbjct 184 SYNSYAIKKKDELERVAKSNR 204
> cel:T05E11.1 rps-5; Ribosomal Protein, Small subunit family
member (rps-5); K02989 small subunit ribosomal protein S5e
Length=210
Score = 292 bits (748), Expect = 4e-79, Method: Compositional matrix adjust.
Identities = 137/199 (68%), Positives = 165/199 (82%), Gaps = 0/199 (0%)
Query 4 AAAAATMAATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQ 63
A AA A + LFG+W+ V VSD+SLVDY+ VK K+A PH+AGR+Q +RFRKA
Sbjct 12 ADAAPATEAPEVALFGKWSLQSVNVSDISLVDYIPVKEKSAKYLPHSAGRFQVRRFRKAA 71
Query 64 CPLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDS 123
CP+VERL NSLMMHGRN+GKKL +RIV+HAF+II L+ +NP+QVLV AV GPREDS
Sbjct 72 CPIVERLANSLMMHGRNNGKKLMTVRIVKHAFEIIYLLTGENPVQVLVNAVINSGPREDS 131
Query 124 TRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESS 183
TRIG AG VRRQAVDV+PLRRVNQAI+L+CTGAR AAFRN+K+++ECLADE++N AK SS
Sbjct 132 TRIGRAGTVRRQAVDVAPLRRVNQAIWLLCTGAREAAFRNVKTIAECLADELINAAKGSS 191
Query 184 NSYAIKKKDEIERVAKANR 202
NSYAIKKKDE+ERVAK+NR
Sbjct 192 NSYAIKKKDELERVAKSNR 210
> sce:YJR123W RPS5; Rps5p; K02989 small subunit ribosomal protein
S5e
Length=225
Score = 266 bits (680), Expect = 3e-71, Method: Compositional matrix adjust.
Identities = 120/192 (62%), Positives = 155/192 (80%), Gaps = 2/192 (1%)
Query 11 AATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLVERL 70
A T+ KLF +W++++V V D SLVDY+ V+ + HTAGRY KRFRKAQCP++ERL
Sbjct 36 AQTEIKLFNKWSFEEVEVKDASLVDYVQVR--QPIFVAHTAGRYANKRFRKAQCPIIERL 93
Query 71 VNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAG 130
NSLMM+GRN+GKKL+A+RI++H DII ++ D+NP+QV+V A+ GPRED+TR+G G
Sbjct 94 TNSLMMNGRNNGKKLKAVRIIKHTLDIINVLTDQNPIQVVVDAITNTGPREDTTRVGGGG 153
Query 131 VVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKK 190
RRQAVDVSPLRRVNQAI L+ GAR AAFRN+K+++E LA+E++N AK SS SYAIKK
Sbjct 154 AARRQAVDVSPLRRVNQAIALLTIGAREAAFRNIKTIAETLAEELINAAKGSSTSYAIKK 213
Query 191 KDEIERVAKANR 202
KDE+ERVAK+NR
Sbjct 214 KDELERVAKSNR 225
> eco:b3341 rpsG, ECK3328, JW3303; 30S ribosomal subunit protein
S7; K02992 small subunit ribosomal protein S7
Length=179
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 46/139 (33%), Positives = 74/139 (53%), Gaps = 13/139 (9%)
Query 66 LVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPRED--S 123
L+ + VN LM+ GKK A IV A + +A + K+ L+ A+E P + S
Sbjct 22 LLAKFVNILMV----DGKKSTAESIVYSALETLAQRSGKSELEAFEVALENVRPTVEVKS 77
Query 124 TRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESS 183
R+G G + V+V P+RR A+ I AR R KSM+ LA+E+ + A+
Sbjct 78 RRVG--GSTYQVPVEVRPVRRNALAMRWIVEAARK---RGDKSMALRLANELSDAAENKG 132
Query 184 NSYAIKKKDEIERVAKANR 202
A+KK++++ R+A+AN+
Sbjct 133 T--AVKKREDVHRMAEANK 149
> ath:ArthCp088 rps7; 30S ribosomal protein S7
Length=155
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 45/139 (32%), Positives = 70/139 (50%), Gaps = 13/139 (9%)
Query 66 LVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGP--REDS 123
LV LVN ++ HG KK A +I+ A I + NPL VL QA+ P +
Sbjct 22 LVNMLVNRILKHG----KKSLAYQIIYRALKKIQQKTETNPLSVLRQAIRGVTPDIAVKA 77
Query 124 TRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESS 183
R+G G + +++ + AI + +R RN M+ L+ E+++ AK S
Sbjct 78 RRVG--GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRN---MAFKLSSELVDAAKGSG 132
Query 184 NSYAIKKKDEIERVAKANR 202
+ AI+KK+E R+A+ANR
Sbjct 133 D--AIRKKEETHRMAEANR 149
> ath:ArthCp069 rps7; 30S ribosomal protein S7
Length=155
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 45/139 (32%), Positives = 70/139 (50%), Gaps = 13/139 (9%)
Query 66 LVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGP--REDS 123
LV LVN ++ HG KK A +I+ A I + NPL VL QA+ P +
Sbjct 22 LVNMLVNRILKHG----KKSLAYQIIYRALKKIQQKTETNPLSVLRQAIRGVTPDIAVKA 77
Query 124 TRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESS 183
R+G G + +++ + AI + +R RN M+ L+ E+++ AK S
Sbjct 78 RRVG--GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRN---MAFKLSSELVDAAKGSG 132
Query 184 NSYAIKKKDEIERVAKANR 202
+ AI+KK+E R+A+ANR
Sbjct 133 D--AIRKKEETHRMAEANR 149
> sce:YJR113C RSM7; Mitochondrial ribosomal protein of the small
subunit, has similarity to E. coli S7 ribosomal protein;
K02989 small subunit ribosomal protein S5e
Length=247
Score = 32.3 bits (72), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 32/136 (23%), Positives = 67/136 (49%), Gaps = 8/136 (5%)
Query 67 VERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRI 126
++ + N +M HG KK +A I+ A ++ ++P+Q L ++++ P T+
Sbjct 114 LDHITNMIMRHG----KKEKAQTILSRALYLVYCQTRQDPIQALEKSLDELAPLM-MTKT 168
Query 127 GSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSY 186
+ GV + + V PL + + ++A R + L +E+ AK +S+++
Sbjct 169 FNTGVAKASVIPV-PLNKRQRNRIAWNWIVQSANQRVSSDFAVRLGEELTAIAKGTSSAF 227
Query 187 AIKKKDEIERVAKANR 202
+K+D+I + A A+R
Sbjct 228 --EKRDQIHKTAIAHR 241
> dre:619204 mrps7, MGC110673, zgc:110673; mitochondrial ribosomal
protein S7
Length=228
Score = 32.0 bits (71), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 38/154 (24%), Positives = 62/154 (40%), Gaps = 24/154 (15%)
Query 65 PLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMA---------------DKNPLQV 109
PL+ + +N +M G K+ A I+ + I + NP +
Sbjct 75 PLISKFINMMMY----DGNKVLARGIMTQTLETIKRKQVEKYHKSPAAKREEIECNPYAI 130
Query 110 LVQAVERGGPREDSTRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGART-AAFRNLKSMS 168
QA+E P I G + V ++ RR A+ + T RT R L M
Sbjct 131 FHQAMENCKPVIGLASIQKGGKFYQVPVPLTDNRRRFMAMKWMITECRTNKQGRTL--MY 188
Query 169 ECLADEIMNCAKESSNSYAIKKKDEIERVAKANR 202
E L+ E++ N IK+K ++ ++A+ANR
Sbjct 189 EKLSQELLAAFANEGN--VIKRKHDLHKMAEANR 220
> dre:559710 tapt1; similar to FLJ90013 protein
Length=567
Score = 31.6 bits (70), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 28 VSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLVERLVNSLMMHG 78
VSDLSLV +++ + H +Y ++R R C + + + LM+ G
Sbjct 62 VSDLSLVRFISAELTRGYFLEHNEAKYTERRERVYTCLRIPKELEKLMIFG 112
> ath:ArthMp103 rps7; ribosomal protein S7
Length=148
Score = 31.2 bits (69), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 35/140 (25%), Positives = 65/140 (46%), Gaps = 7/140 (5%)
Query 63 QCPLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPRED 122
Q L+++LVN M GK+ + IV F A +++ ++++V AVE P +
Sbjct 8 QKLLIKKLVNFRM----KEGKRTRVRAIVYQTFHRPA-RTERDVIKLMVDAVENIKPICE 62
Query 123 STRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKES 182
++G AG + V+ R+ AI I A S+ +C EI++ ++
Sbjct 63 VAKVGVAGTIYDVPGIVARDRQQTLAIRWILEAAFKRRISYRISLEKCSFAEILDAYQKR 122
Query 183 SNSYAIKKKDEIERVAKANR 202
+ A +K++ + +A NR
Sbjct 123 GS--ARRKRENLHGLASTNR 140
> ath:AT2G07696 ribosomal protein S7 family protein
Length=148
Score = 31.2 bits (69), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 35/140 (25%), Positives = 65/140 (46%), Gaps = 7/140 (5%)
Query 63 QCPLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPRED 122
Q L+++LVN M GK+ + IV F A +++ ++++V AVE P +
Sbjct 8 QKLLIKKLVNFRM----KEGKRTRVRAIVYQTFHRPA-RTERDVIKLMVDAVENIKPICE 62
Query 123 STRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKES 182
++G AG + V+ R+ AI I A S+ +C EI++ ++
Sbjct 63 VAKVGVAGTIYDVPGIVARDRQQTLAIRWILEAAFKRRISYRISLEKCSFAEILDAYQKR 122
Query 183 SNSYAIKKKDEIERVAKANR 202
+ A +K++ + +A NR
Sbjct 123 GS--ARRKRENLHGLASTNR 140
> dre:560056 znf704, si:ch211-203l9., si:ch211-203l9.2, si:ch211-203l9.3;
zinc finger protein 704
Length=501
Score = 30.8 bits (68), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 33/57 (57%), Gaps = 0/57 (0%)
Query 85 LQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAGVVRRQAVDVSP 141
+Q + +++ + IA M K PL+ +++ GG R+ T+ + GV R +A+ +SP
Sbjct 55 MQDGSVKEYSEEEIAQMTAKAPLKSSLKSTCNGGQRDGLTQGQNGGVQRGEALSLSP 111
> cpv:cgd5_1280 tfc4p like TFIIIC subunit; TPR repeat containing
basal transcription factor
Length=933
Score = 30.0 bits (66), Expect = 6.1, Method: Composition-based stats.
Identities = 20/73 (27%), Positives = 35/73 (47%), Gaps = 8/73 (10%)
Query 129 AGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAI 188
A V+ +D+ +R+NQ IY +R N K+ EC+ +I + + S NS
Sbjct 558 ANVINEFIIDI---KRINQLIY-----SRKYNENNKKNSLECIEKDISSTSNYSKNSNET 609
Query 189 KKKDEIERVAKAN 201
+ +R+ +AN
Sbjct 610 STLNTSKRLQRAN 622
Lambda K H
0.320 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6124680020
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40