bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2940_orf1
Length=330
Score E
Sequences producing significant alignments: (Bits) Value
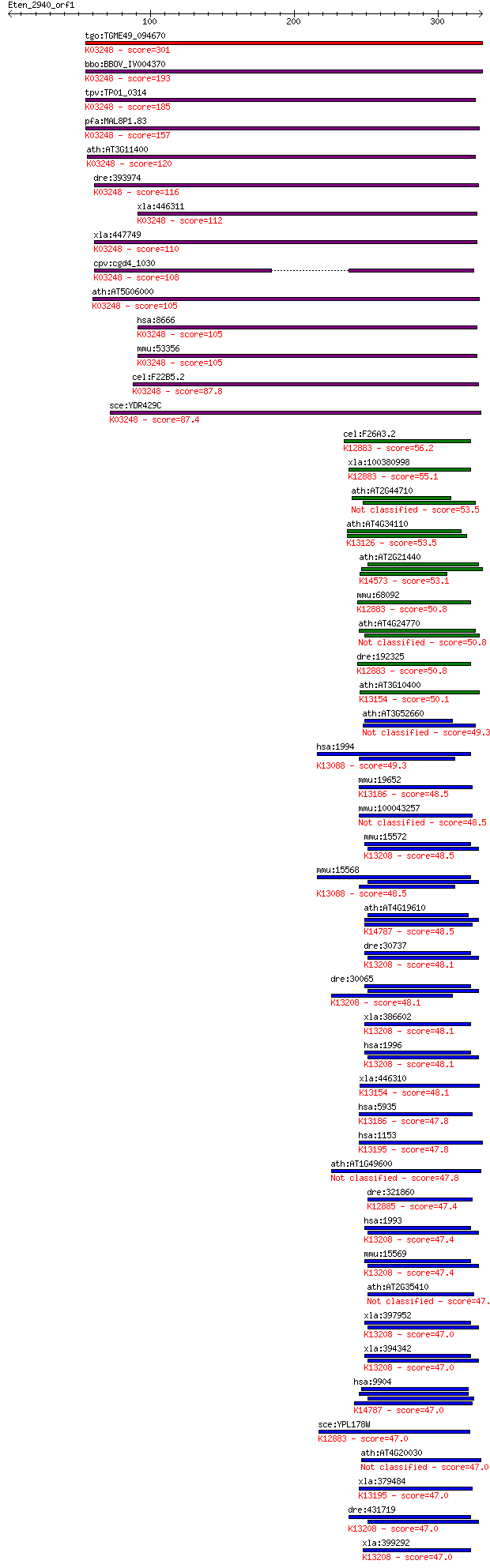
tgo:TGME49_094670 eukaryotic translation initiation factor 3, ... 301 3e-81
bbo:BBOV_IV004370 23.m06488; translation initiation factor 3 s... 193 9e-49
tpv:TP01_0314 eukaryotic translation initiation factor 3 subun... 185 3e-46
pfa:MAL8P1.83 eukaryotictranslation initiation factor, putativ... 157 4e-38
ath:AT3G11400 EIF3G1; eukaryotic translation initiation factor... 120 1e-26
dre:393974 eif3g, MGC56553, eif3s4, zgc:56553; eukaryotic tran... 116 1e-25
xla:446311 eif3s4, MGC83369, eIF3g-A, eif3s4-A; eukaryotic tra... 112 2e-24
xla:447749 eIF3g-B, eif3s4-B; MGC83338 protein; K03248 transla... 110 1e-23
cpv:cgd4_1030 eukaryotic translation initiation factor ; K0324... 108 2e-23
ath:AT5G06000 EIF3G2; EIF3G2; RNA binding / translation initia... 105 2e-22
hsa:8666 EIF3G, EIF3-P42, EIF3S4, eIF3-delta, eIF3-p44; eukary... 105 2e-22
mmu:53356 Eif3g, 44kDa, D0Jmb4, Eif3s4, TU-189B2, eIF3-p44, p4... 105 3e-22
cel:F22B5.2 eif-3.G; Eukaryotic Initiation Factor family membe... 87.8 4e-17
sce:YDR429C TIF35; Tif35p; K03248 translation initiation facto... 87.4 6e-17
cel:F26A3.2 ncbp-2; Nuclear Cap-Binding Protein family member ... 56.2 1e-07
xla:100380998 ncbp2; nuclear cap binding protein subunit 2, 20... 55.1 3e-07
ath:AT2G44710 RNA recognition motif (RRM)-containing protein 53.5 1e-06
ath:AT4G34110 PAB2; PAB2 (POLY(A) BINDING 2); RNA binding / tr... 53.5 1e-06
ath:AT2G21440 RNA recognition motif (RRM)-containing protein; ... 53.1 2e-06
mmu:68092 Ncbp2, 20kDa, 5930413E18Rik, AA536802, AI843301, C79... 50.8 7e-06
ath:AT4G24770 RBP31; RBP31 (31-KDA RNA BINDING PROTEIN); RNA b... 50.8 7e-06
dre:192325 ncbp2, CHUNP6917, zgc:92592; nuclear cap binding pr... 50.8 7e-06
ath:AT3G10400 RNA recognition motif (RRM)-containing protein; ... 50.1 1e-05
ath:AT3G52660 RNA recognition motif (RRM)-containing protein 49.3 2e-05
hsa:1994 ELAVL1, ELAV1, HUR, Hua, MelG; ELAV (embryonic lethal... 49.3 2e-05
mmu:19652 Rbm3, 2600016C11Rik, MGC118410; RNA binding motif pr... 48.5 3e-05
mmu:100043257 Gm15453, OTTMUSG00000022091; predicted gene 15453 48.5
mmu:15572 Elavl4, Elav, Hud, PNEM; ELAV (embryonic lethal, abn... 48.5 3e-05
mmu:15568 Elavl1, 2410055N02Rik, HUR, Hua, W91709; ELAV (embry... 48.5 3e-05
ath:AT4G19610 RNA binding / nucleic acid binding / nucleotide ... 48.5 4e-05
dre:30737 elavl4, elrd, wu:fc24h01, zHuD; ELAV (embryonic leth... 48.1 4e-05
dre:30065 hug, fb72d01, fk65g02, wu:fb72d01, wu:fk65g02, zHuG;... 48.1 4e-05
xla:386602 elavl4, HuD, elrD, elrD1, elrD2; ELAV (embryonic le... 48.1 4e-05
hsa:1996 ELAVL4, HUD, PNEM; ELAV (embryonic lethal, abnormal v... 48.1 4e-05
xla:446310 zcrb1, MGC82154; zinc finger CCHC-type and RNA bind... 48.1 5e-05
hsa:5935 RBM3, IS1-RNPL, RNPL; RNA binding motif (RNP1, RRM) p... 47.8 6e-05
hsa:1153 CIRBP, CIRP; cold inducible RNA binding protein; K131... 47.8 6e-05
ath:AT1G49600 ATRBP47A (Arabidopsis thaliana RNA-binding prote... 47.8 7e-05
dre:321860 rbmx, fc66e11, wu:fb37c07, wu:fc66e11; RNA binding ... 47.4 7e-05
hsa:1993 ELAVL2, HEL-N1, HELN1, HUB; ELAV (embryonic lethal, a... 47.4 7e-05
mmu:15569 Elavl2, Hub, mel-N1; ELAV (embryonic lethal, abnorma... 47.4 8e-05
ath:AT2G35410 33 kDa ribonucleoprotein, chloroplast, putative ... 47.4 9e-05
xla:397952 elavl2-b, elavl2, elrB, hel-n1, heln1, hub, xel-1; ... 47.0 9e-05
xla:394342 elavl2-a, Xel-1, el-1, elavl2, elrB, hel-n1, heln1,... 47.0 9e-05
hsa:9904 RBM19, DKFZp586F1023, KIAA0682; RNA binding motif pro... 47.0 9e-05
sce:YPL178W CBC2, CBP20, MUD13, SAE1; Cbc2p; K12883 nuclear ca... 47.0 9e-05
ath:AT4G20030 RNA recognition motif (RRM)-containing protein 47.0 9e-05
xla:379484 cirbp-b, MGC64461, Xcirp, cirbp, cirp, cirp2, xcirp... 47.0 1e-04
dre:431719 elavl2, zgc:91918; ELAV (embryonic lethal, abnormal... 47.0 1e-04
xla:399292 elavl1-a, Elavl1, MGC132161, Xel-1, elav1, elrA, hu... 47.0 1e-04
> tgo:TGME49_094670 eukaryotic translation initiation factor 3,
putative ; K03248 translation initiation factor 3 subunit
G
Length=277
Score = 301 bits (770), Expect = 3e-81, Method: Compositional matrix adjust.
Identities = 163/278 (58%), Positives = 203/278 (73%), Gaps = 3/278 (1%)
Query 55 MTTDRKWADFELEDDGDLGDLAPTPGGFETRPDSDGIKTVTNYLQNAKGETLKIVKRIKV 114
M +RKWADFEL+DD +LGDL T GFET+PD +G+KTVT Y Q +GETLK+ KRIKV
Sbjct 1 MAAERKWADFELDDDYELGDLVNTATGFETKPDEEGVKTVTTYTQTLRGETLKVTKRIKV 60
Query 115 TRVVHRINKAVHERRNLPLFGIEAAEGSTG-KAFTVRSVEEIPLEISKETLLRAKFKEDE 173
R RINK V+ R+N+ FG++AA+G A TVRS EEI +E+ + + R +E+E
Sbjct 61 IRKCQRINKDVYARKNIVPFGLDAADGDPSLSATTVRSHEEIIIEVPRSSKQRKFKEEEE 120
Query 174 DDEDIQFADLNPAKTSRDLRQKFLSLQNDYEPVDAG-EGDPLGSRLGVPGAKGLSRLRGG 232
+D+ DLNP+KTSRDLRQKF +L+ D E AG +GD K + R R
Sbjct 121 EDDFY-MGDLNPSKTSRDLRQKFRALREDDEAGAAGGDGDEETGLRSAGAGKYIPRARRE 179
Query 233 GDALKDLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAF 292
G+ LKD+E RRREE TIRVTNLSED K+EDL ELFG IGK++R++LAKHK+KK SKGFAF
Sbjct 180 GETLKDVENRRREECTIRVTNLSEDVKDEDLTELFGKIGKIDRIYLAKHKEKKCSKGFAF 239
Query 293 ITYANRELAAQAIRKLNRHGYDNLLLNVEWARPNNRER 330
ITY RE A +AIR+LNRHGYDNLLLNVEWA+P+NR+R
Sbjct 240 ITYQRREDAVRAIRQLNRHGYDNLLLNVEWAKPSNRDR 277
> bbo:BBOV_IV004370 23.m06488; translation initiation factor 3
subunit; K03248 translation initiation factor 3 subunit G
Length=268
Score = 193 bits (490), Expect = 9e-49, Method: Compositional matrix adjust.
Identities = 120/279 (43%), Positives = 173/279 (62%), Gaps = 21/279 (7%)
Query 55 MTTDRKWADFELEDDGDLGDL-APTPGGFETRPDSDGIKTVTNYLQNAKGETLKIVKRIK 113
M ++ KWAD E ++D D+ + + FET PDS GIK VT+Y +N +G+T+KI KRIK
Sbjct 6 MISETKWADIEADEDYDVDGIDSHRLTAFETAPDSQGIKVVTSYTKNRQGQTVKITKRIK 65
Query 114 VTRVVHRINKAVHERRNLPLFGIEAAEGSTGKAFTVRSVEEIPLEISKETLLRAKFKEDE 173
+R+ RI+K R + +F ++ + T+ S EEI +E + + R E
Sbjct 66 ESRIPRRIHKEAKARADGSIFKVDK---NMEAGITMASQEEILIE---QPVGRRSKVNVE 119
Query 174 DDEDIQFA--DLNPAKTSRDLRQKFLSLQNDYEPVDAGEGDPLGSRLGVPGAKGLSRLRG 231
D+ D+ +A D+N + +R+L+ KF SL++D + VD + + ++ P SR G
Sbjct 120 DNLDLVYAPADINLTRATRELKMKFKSLRDDSDVVDMMDSKDMPAKYVPP-----SRKEG 174
Query 232 GGDALKDLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFA 291
G + R +E T+RVTNLSED +E+DL ELF +G++ R +LAKHK+ + SKGFA
Sbjct 175 G-------DRRNFDENTVRVTNLSEDVREKDLVELFSRVGRIHRAYLAKHKETQYSKGFA 227
Query 292 FITYANRELAAQAIRKLNRHGYDNLLLNVEWARPNNRER 330
FITYA R+ A AI KLNR GYDNLLLNVEWA+P NR+R
Sbjct 228 FITYATRQDALNAINKLNRQGYDNLLLNVEWAKPPNRDR 266
> tpv:TP01_0314 eukaryotic translation initiation factor 3 subunit
4; K03248 translation initiation factor 3 subunit G
Length=266
Score = 185 bits (469), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 119/274 (43%), Positives = 168/274 (61%), Gaps = 23/274 (8%)
Query 55 MTTDRKWADFELEDDGDLGDL-APTPGGFETRPDSDGIKTVTNYLQNAKGETLKIVKRIK 113
++++ KWAD E +DD D +L FE DS GIK++ +Y +N++G ++KI KR+K
Sbjct 9 LSSETKWADIEADDDYDNENLNVQHLKEFEIATDSQGIKSLVSYSRNSRGHSVKITKRVK 68
Query 114 VTRVVHRINKAVHERRNLPLFGIEAAEGSTGKAFTVRSVEEIPLEISKETLLRAKFKEDE 173
RV RI+K+V ER+NL F + E TG +F S EEI +E+S+ + +D
Sbjct 69 EIRVSKRIHKSVFERKNLVPFNL-TDENDTGSSFV--SNEEIIIELSRN---ERRLNQD- 121
Query 174 DDEDIQFA--DLNPAKTSRDLRQKFLSLQNDYEPVDAGEGDPLGSRLGVPGAKGLSRLRG 231
DD D+ ++ D N K SRDL+ KF SL+ + + E + VP SR G
Sbjct 122 DDSDLIYSPVDANFLKLSRDLKLKFKSLKEEDNVPEEVEEEAPAKY--VPP----SRKEG 175
Query 232 GGDALKDLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFA 291
G E R ++ TIR+TNLSED +E+DL ELFG +G++ R +LAK+K+ + KGFA
Sbjct 176 G-------ERRSFDDNTIRITNLSEDIREKDLTELFGRVGRIHRAYLAKYKETQNPKGFA 228
Query 292 FITYANRELAAQAIRKLNRHGYDNLLLNVEWARP 325
F+TY N+E A +AI K NR GY+NLLLNVEWARP
Sbjct 229 FVTYVNKEDAREAINKFNRWGYNNLLLNVEWARP 262
> pfa:MAL8P1.83 eukaryotictranslation initiation factor, putative;
K03248 translation initiation factor 3 subunit G
Length=266
Score = 157 bits (398), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 107/292 (36%), Positives = 156/292 (53%), Gaps = 45/292 (15%)
Query 55 MTTDRKWADFELEDDGDLGDLAPTPGGFETRPDSD---------------GIKTVTNYLQ 99
+ T++KWAD +++DD + D + D D GIK VT Y +
Sbjct 2 IQTEKKWADIDVDDDESINDNEIKKKWEDIDADDDLEANKKLSYFTNYENGIKVVTKYSE 61
Query 100 NAKGETLKIVKRIKVTRVVHRINKAVHERRNLPLFGIEAAEGSTGKAFTVRSVE---EIP 156
N K +T + K+I+ + ++NK ++ R NL F I+ T + V ++E P
Sbjct 62 NLKKQT--VTKKIQEVVIKKKLNKEINNRLNLKNFNIDIHSSVTVEPTDVVNIEVPKNNP 119
Query 157 LEISKETLLRAKFKEDEDDEDIQFADLNPAKTSRDLRQKFLSLQNDYEPVDAGEGDPLGS 216
L+ K+T + D FA+ KT++DL+ +F L+ D E D DP
Sbjct 120 LDFLKDT-----------EYDYLFAE-QTNKTAKDLKNRFKILK-DEETEDVKAEDP--- 163
Query 217 RLGVPGAKGLSRLRGGGDALKDLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERV 276
+K R G D + + E T+RVTNLSED E +L+ LFG +G + R+
Sbjct 164 ------SKAAGRREGLYYRPHDTQNK---ECTVRVTNLSEDVNENELSNLFGKVGNIVRM 214
Query 277 FLAKHKDKKASKGFAFITYANRELAAQAIRKLNRHGYDNLLLNVEWARPNNR 328
FLAKHK+ + SKGFAFITY+ RE A +AI KLNRHG++NLLL+VEWA+P+NR
Sbjct 215 FLAKHKETQNSKGFAFITYSKREEAKRAIEKLNRHGFENLLLSVEWAKPSNR 266
> ath:AT3G11400 EIF3G1; eukaryotic translation initiation factor
3G / eIF3g; K03248 translation initiation factor 3 subunit
G
Length=321
Score = 120 bits (300), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 103/315 (32%), Positives = 144/315 (45%), Gaps = 50/315 (15%)
Query 56 TTDRKWADFELEDDGDLGDLAPTPGGFETRPDSDGIKTVTNYLQNAKGETLKIVKRIKVT 115
T+ +W E+++D DL L P PD +G+KT Y N + +KI R +V
Sbjct 9 TSKFRWG--EMDEDEDLDFLLPPKQVIG--PDENGLKTTIEYKFNDEENKVKITTRTRVR 64
Query 116 RVVH-RINKAVHERRNLPLFGIEAAEGSTGKAFTVRSVEEIPLEISKE------TLLRAK 168
++ R+NK ERRN P FG +AA G T+ S EEI LE + + A
Sbjct 65 KLASARLNKRAMERRNWPKFG-DAANEEAGSHLTMVSTEEILLERPRAPGDQLANHMLAS 123
Query 169 FKEDEDDEDIQFADLNPAK------TSRDLRQ-----------KFLSLQNDY-------- 203
E E + F N K T L Q + + D+
Sbjct 124 IPEYELKCALTFVAANCTKADESKATGDGLSQLGKGGAVLMVCRICHKKGDHWTSKCPYK 183
Query 204 ---EPVDAG-EGDPLGSRLGVPGAKGLSR-------LRGGGD--ALKDLEARRREEATIR 250
P D + P G + A G + +R G D A+ RR +E ++R
Sbjct 184 DLAAPTDVFIDKPPTGESSTMSAAPGTGKAAYVPPSMRAGADRSAVGSDMRRRNDENSVR 243
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
VTNLSED +E DL ELF G + RV++A + S+GF F+ + +RE A +AI KLN
Sbjct 244 VTNLSEDTREPDLMELFHPFGAVTRVYVAIDQKTGVSRGFGFVNFVSREDAQRAINKLNG 303
Query 311 HGYDNLLLNVEWARP 325
+GYDNL+L VEWA P
Sbjct 304 YGYDNLILRVEWATP 318
> dre:393974 eif3g, MGC56553, eif3s4, zgc:56553; eukaryotic translation
initiation factor 3, subunit G; K03248 translation
initiation factor 3 subunit G
Length=293
Score = 116 bits (291), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 100/291 (34%), Positives = 147/291 (50%), Gaps = 35/291 (12%)
Query 61 WADFELEDDGDLGDLAPTPGGFETRPDSDGIKTVTNYLQNAKGETLKIVKRIKVTRVVHR 120
WAD ++E++GD G L P+P ET IKTVT Y + G+ KI++ K+ +
Sbjct 13 WAD-QVEEEGDEGSL-PSPK--ETV--KGNIKTVTEYKIDDDGQKFKIIRTFKIE--TRK 64
Query 121 INKAVHERRNLPLFGIEAAEGSTGKAFTVRSVEEIPLE-ISKETLLRAKFKEDEDDEDI- 178
+KAV R+N FG + T +++ + IS + L A +D+D++ +
Sbjct 65 ASKAVARRKNWKKFGNSEYDAPGPNVATTTVSDDVFMTFISSKEDLNA---QDQDEDPMN 121
Query 179 ----------------QFADLNPAKTSRDLRQKFLSLQ---NDYEPVDAGEGDPLGSRLG 219
+ P K + QK L+ Q + E A E +P
Sbjct 122 KLEGQKIVSCRICKGDHWTTRCPYKDTLGPMQKELAEQLGLSTGEKEKAAEPEPAQPVQN 181
Query 220 VPGAKGLSRLRGGGDAL-KDLEARRR--EEATIRVTNLSEDAKEEDLAELFGNIGKLERV 276
G LR GG + ++ RR + ATIRVTNLSED +E DL ELF G + R+
Sbjct 182 KTGKYVPPSLRDGGTRRGESMQPNRRADDNATIRVTNLSEDTRETDLQELFRPFGSISRI 241
Query 277 FLAKHKDKKASKGFAFITYANRELAAQAIRKLNRHGYDNLLLNVEWARPNN 327
+LAK K+ SKGFA I++ RE AA+AI ++ GYD+L+LNVEWA+P+N
Sbjct 242 YLAKDKNTGQSKGFALISFHRREDAARAIAGVSGFGYDHLILNVEWAKPSN 292
> xla:446311 eif3s4, MGC83369, eIF3g-A, eif3s4-A; eukaryotic translation
initiation factor 3, subunit 4 delta, 44kDa; K03248
translation initiation factor 3 subunit G
Length=308
Score = 112 bits (279), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 87/258 (33%), Positives = 130/258 (50%), Gaps = 24/258 (9%)
Query 91 IKTVTNYLQNAKGETLKIVKRIKVTRVVHRINKAVHERRNLPLFGIEAAEGSTGKAFTVR 150
IKT+T Y N +T+KIV+ K+ V + +K V +R+N FG + T
Sbjct 51 IKTITEYKLNDDDKTVKIVRTFKIETV--KASKVVAQRKNWKKFGNSEYDPPGPNVATTT 108
Query 151 SVEEIPLE-ISKETLLRAKFKEDEDDE-------------DIQFADLNPAKTSRDLRQKF 196
+++ + I+ + L + +ED ++ + P K + QK
Sbjct 109 VSDDVLMTFITNKEDLNNQEEEDPMNKLKGQKIVSCRICKGDHWTTRCPYKDTLGPMQKE 168
Query 197 LSLQ-----NDYEPVDAGEGDPLGSRLGVPGAKGLSRLRGGGDAL-KDLEARRR--EEAT 248
L+ Q D E E +P + + G LR GG + ++ RR + AT
Sbjct 169 LAEQLGLSTGDKEKAPGAEPEPAQAPVSKTGKYVPPSLRDGGSRRGESMQPNRRADDNAT 228
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
IRVTNLSED +E DL ELF G + R++LAK K SKGFAFI++ RE AA+AI +
Sbjct 229 IRVTNLSEDTRETDLQELFRPFGSISRIYLAKDKTTGQSKGFAFISFHRREDAARAIAGV 288
Query 309 NRHGYDNLLLNVEWARPN 326
+ GYD+L+LNVEWA+P+
Sbjct 289 SGFGYDHLILNVEWAKPS 306
> xla:447749 eIF3g-B, eif3s4-B; MGC83338 protein; K03248 translation
initiation factor 3 subunit G
Length=308
Score = 110 bits (274), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 97/297 (32%), Positives = 144/297 (48%), Gaps = 33/297 (11%)
Query 61 WADFELEDDGDLGDLAPT-----PGGF---ETRPDSDG-IKTVTNYLQNAKGETLKIVKR 111
WAD E+ D+ L+P P F R +G IKT+T Y N K + +KIV+
Sbjct 12 WADQVEEEGIDVEPLSPQIKKQDPASFVLDTPREVINGNIKTITEYKLNDKDKKIKIVRT 71
Query 112 IKVTRVVHRINKAVHERRNLPLFGIEAAEGSTGKAFTVRSVEEIPLE-ISKETLLRAKFK 170
K+ + + +K V R+N FG + T +++ + I+ + L + +
Sbjct 72 FKIETL--KASKVVAHRKNWKKFGNSEYDPPGPNVATTTVSDDVLMTFITNKEDLNNQEE 129
Query 171 EDEDDE-------------DIQFADLNPAKTSRDLRQKFLSLQ-----NDYEPVDAGEGD 212
ED ++ + P K + QK L+ Q D E E +
Sbjct 130 EDPMNKLKGQKIVSCRICKGDHWTTRCPYKDTLGPMQKELAEQLGLSTADKEKAPGAEPE 189
Query 213 PLGSRLGVPGAKGLSRLRGGGDAL-KDLEARRR--EEATIRVTNLSEDAKEEDLAELFGN 269
P + + G LR GG + ++ RR + ATIRVTNLSED +E DL ELF
Sbjct 190 PAQAPVSKTGKYVPPSLRDGGSRRGESMQPNRRADDNATIRVTNLSEDTRETDLQELFRP 249
Query 270 IGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNRHGYDNLLLNVEWARPN 326
G + R++LAK K SKGFAFI++ RE AA+AI ++ GYD+L+LNVEWA+P+
Sbjct 250 FGSISRIYLAKDKTTGQSKGFAFISFHRREDAARAIAGVSGFGYDHLILNVEWAKPS 306
> cpv:cgd4_1030 eukaryotic translation initiation factor ; K03248
translation initiation factor 3 subunit G
Length=352
Score = 108 bits (271), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 55/88 (62%), Positives = 69/88 (78%), Gaps = 1/88 (1%)
Query 238 DLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASK-GFAFITYA 296
+L++ R++ T+RV NLSEDA EEDL ELF G++ +VFLAK+K+ + GFAFITY+
Sbjct 261 NLQSSHRDDCTVRVANLSEDATEEDLQELFKTAGRVVKVFLAKNKNNSKNTKGFAFITYS 320
Query 297 NRELAAQAIRKLNRHGYDNLLLNVEWAR 324
RE A AIRKLNRHGYDNLLLNVEWA+
Sbjct 321 KREEAQNAIRKLNRHGYDNLLLNVEWAK 348
Score = 64.3 bits (155), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 44/127 (34%), Positives = 68/127 (53%), Gaps = 13/127 (10%)
Query 61 WADFELEDDGDLGDLAPT----PGGFETRPDSDGIKTVTNYLQNAKGETLKIVKRIKVTR 116
WAD D+ D P+ GFE++PD DGIK V +Y++++KG T+KI K+IK R
Sbjct 13 WAD------EDIDDFEPSFDEPLKGFESKPDKDGIKHVISYIKDSKGSTIKITKKIKEVR 66
Query 117 VVHRINKAVHERRNLPLFGIEAAEGSTGKAFTVRSVEEIPLEISKETLLRAKFKEDEDDE 176
+ +INK V RR + A+ + +EI ++I K T F++ ++D+
Sbjct 67 NIIKINKNVVNRR--LIMERNLADRFNSDTDVPKLGDEISIDIPKNT-DSMNFQDKDNDD 123
Query 177 DIQFADL 183
D F DL
Sbjct 124 DYYFMDL 130
> ath:AT5G06000 EIF3G2; EIF3G2; RNA binding / translation initiation
factor; K03248 translation initiation factor 3 subunit
G
Length=308
Score = 105 bits (263), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 96/289 (33%), Positives = 139/289 (48%), Gaps = 34/289 (11%)
Query 60 KWADFELEDDGDLGDLAPTPGGFETRPDSDGIKTVTNYLQNAKGETLKIVKRIKVTRVVH 119
+W + + E++GD L P PD +G+K V Y N + + +KI TRV
Sbjct 13 RWGEID-EEEGDYDFLLPPKQMIS--PDQNGVKKVIEYKFNEEDKKVKITT---TTRVQK 66
Query 120 R-INKAVHERRNLPLFGIEAAEGSTGKAFTVRSVEEIPLEISKETLLRAKFKEDEDDEDI 178
R + K ERR+ FG +AA + T+RS E+I LE + A+ D
Sbjct 67 RALTKQAVERRSWNKFG-DAAHEESSSYLTMRSTEDIILERIRAPGSNAEQSTVSGDSMS 125
Query 179 QF----ADLNPAK---------TSRDLRQKFLSLQNDYEPVDAGEGDPLGSRLG----VP 221
Q A L + TSR ++ LSL + EP+ A + G VP
Sbjct 126 QLGKPGAVLMVCRLCQKKGDHWTSRCPQKDLLSLMD--EPLTAETSTSTITGTGRAAYVP 183
Query 222 GA--KGLSRLRGGGDALKDLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLA 279
+ +G R GG D R +E ++RVTNLSED + DL ELF G + R +A
Sbjct 184 PSMREGADRKAGGSDMRS-----RHDENSVRVTNLSEDTRGPDLMELFRPFGAVTRCHVA 238
Query 280 KHKDKKASKGFAFITYANRELAAQAIRKLNRHGYDNLLLNVEWARPNNR 328
+ S+GF F+++ +RE A +AI KLN +GYDNL+L VEW+ P +
Sbjct 239 IDQKTSMSRGFGFVSFVSREDAQRAINKLNGYGYDNLILRVEWSTPKTQ 287
> hsa:8666 EIF3G, EIF3-P42, EIF3S4, eIF3-delta, eIF3-p44; eukaryotic
translation initiation factor 3, subunit G; K03248 translation
initiation factor 3 subunit G
Length=320
Score = 105 bits (262), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 89/260 (34%), Positives = 126/260 (48%), Gaps = 29/260 (11%)
Query 91 IKTVTNYLQNAKGETLKIVKRIKVTRVVHRINKAVHERRNLPLFGIEAAE--GSTGKAFT 148
IKTVT Y + G+ KIV+ ++ + +KAV R+N FG + G T
Sbjct 64 IKTVTEYKIDEDGKKFKIVRTFRIE--TRKASKAVARRKNWKKFGNSEFDPPGPNVATTT 121
Query 149 VRSVEEIPLEISKETLLRAKFKEDEDDEDI---------------QFADLNPAKTSRDLR 193
V + SKE L +E+ED + + P K +
Sbjct 122 VSDDVSMTFITSKEDL---NCQEEEDPMNKLKGQKIVSCRICKGDHWTTRCPYKDTLGPM 178
Query 194 QKFLSLQNDYEPVD----AGEGDPLGSRLGVPGAKGLSRLRGGGDAL-KDLEARRR--EE 246
QK L+ Q + GE +P+ + G LR G + ++ RR +
Sbjct 179 QKELAEQLGLSTGEKEKLPGELEPVQATQNKTGKYVPPSLRDGASRRGESMQPNRRADDN 238
Query 247 ATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIR 306
ATIRVTNLSED +E DL ELF G + R++LAK K SKGFAFI++ RE AA+AI
Sbjct 239 ATIRVTNLSEDTRETDLQELFRPFGSISRIYLAKDKTTGQSKGFAFISFHRREDAARAIA 298
Query 307 KLNRHGYDNLLLNVEWARPN 326
++ GYD+L+LNVEWA+P+
Sbjct 299 GVSGFGYDHLILNVEWAKPS 318
> mmu:53356 Eif3g, 44kDa, D0Jmb4, Eif3s4, TU-189B2, eIF3-p44,
p44; eukaryotic translation initiation factor 3, subunit G;
K03248 translation initiation factor 3 subunit G
Length=320
Score = 105 bits (261), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 95/271 (35%), Positives = 131/271 (48%), Gaps = 51/271 (18%)
Query 91 IKTVTNYLQNAKGETLKIVKRIKVTRVVHRINKAVHERRNLPLFGIEAAE--GSTGKAFT 148
IKTVT Y G+ KIV+ ++ + +KAV R+N FG + G T
Sbjct 64 IKTVTEYKIEEDGKKFKIVRTFRIE--TRKASKAVARRKNWKKFGNSEFDPPGPNVATTT 121
Query 149 VRSVEEIPLEISKETLLRAKFKEDEDDEDI---------------QFADLNPAKTSRDLR 193
V + SKE L +E+ED + + P K +
Sbjct 122 VSDDVSMTFITSKEDL---NCQEEEDPMNKLKGQKIVSCRICKGDHWTTRCPYKDTLGPM 178
Query 194 QKFLS------------LQNDYEPVDAGEGDPLGSRLG--VPGA--KGLSRLRGGGDALK 237
QK L+ L + EPV A + S+ G VP + G SR RG +
Sbjct 179 QKELAEQLGLSTGEKEKLPGELEPVQAAQ-----SKTGKYVPPSLRDGASR-RG-----E 227
Query 238 DLEARRR--EEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITY 295
++ RR + ATIRVTNLSED +E DL ELF G + R++LAK K SKGFAFI++
Sbjct 228 SMQPNRRADDNATIRVTNLSEDTRETDLQELFRPFGSISRIYLAKDKTTGQSKGFAFISF 287
Query 296 ANRELAAQAIRKLNRHGYDNLLLNVEWARPN 326
RE AA+AI ++ GYD+L+LNVEWA+P+
Sbjct 288 HRREDAARAIAGVSGFGYDHLILNVEWAKPS 318
> cel:F22B5.2 eif-3.G; Eukaryotic Initiation Factor family member
(eif-3.G); K03248 translation initiation factor 3 subunit
G
Length=256
Score = 87.8 bits (216), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 74/248 (29%), Positives = 123/248 (49%), Gaps = 25/248 (10%)
Query 88 SDGIKTVTNYLQNAKGETLKIVKRIKVTRVVHRINKAVHERRNLPLFGIEAAEGSTGKAF 147
+DG +T T + + G K+V + KV + R+ K V +R+ FG E + +
Sbjct 26 ADGTRTETAFTE-VDGVRWKVVTQFKV--INKRVPKVVADRKKWVKFGSCKGEPAGPQVA 82
Query 148 TVRSVEEIPLEISKETLLRAKFKEDEDDEDIQFADLNPAKTSRDLR-QKFLSLQNDY--- 203
T EE+ ++ ++ RA + + ED Q A + R + S Y
Sbjct 83 TTYVAEEVDMQFTRN---RAGEQILDVQEDKQTAKTTSREHCRHCKGNDHWSTHCPYKVM 139
Query 204 ----EPVDAGEGDPLGSRLGVPGAKGLSRLRGGGDALKDLEARRREEATIRVTNLSEDAK 259
E DA + D R+ + +R G + ++ R +E T RVTNL ++
Sbjct 140 YQLDEEADA-DKDTEKDRMAMG-------MRPDG---RQIDRNRSDENTCRVTNLPQEMN 188
Query 260 EEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNRHGYDNLLLN 319
E++L +LFG IG++ R+F+A+ K KGFAF+T+ +R+ AA+AI +LN +++L
Sbjct 189 EDELRDLFGKIGRVIRIFIARDKVTGLPKGFAFVTFESRDDAARAIAELNDIRMYHMVLK 248
Query 320 VEWARPNN 327
VEW RP+N
Sbjct 249 VEWTRPSN 256
> sce:YDR429C TIF35; Tif35p; K03248 translation initiation factor
3 subunit G
Length=274
Score = 87.4 bits (215), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 79/288 (27%), Positives = 138/288 (47%), Gaps = 44/288 (15%)
Query 72 LGDLAPTPGGFETRPDSDGIKTVTNYLQNAKGETLKIVKRIKVTRVVHRINKAVHERRNL 131
+ ++AP E ++DG +++ Y + G KI +++K +V+ +++K+V ER+N
Sbjct 1 MSEVAPE----EIIENADGSRSIITY-KIEDGVKYKITQKVKEVKVLEKVHKSVAERKNW 55
Query 132 PLFGIEAAEGSTGKAFTVRSVEEIPLEISKETLLRAKFKEDEDDEDIQFADLNPAKTSRD 191
+G E + A T R EE+ L +S+ +K+ E+ E IQ + KT
Sbjct 56 HKYGSEKGSPAGPSAVTARLGEEVELRLSR------NWKQAEE-ERIQKEKASLTKTGLQ 108
Query 192 LR-------------QKFLSLQNDYEPVDAGEGD----------PLGSRLGVPGAKGLSR 228
R + LS + E EG +G +PG
Sbjct 109 CRLCGNDHMTMNCPFKTILSELSALEDPATNEGGVEAASEEKAGQVGGAGSIPGQYVPPS 168
Query 229 LRGGG-----DALKDLEARRREE-ATIRVTNLSEDAKEEDL-AELFGNIGKLERVFLAKH 281
R G DA +D +R R++ T+++ ++E+A E L EL + RV + ++
Sbjct 169 RRAGARDPSSDAYRD--SRERDDMCTLKIMQVNENADENSLREELLFPFAPIPRVSVVRN 226
Query 282 KDKKASKGFAFITYANRELAAQAIRKLNRHGYDNLLLNVEWARPNNRE 329
K+ S+G AF+T+++ E+A QA+R L+ GY NL+L VEW++P +E
Sbjct 227 KETGKSRGLAFVTFSSEEVAEQALRFLDGRGYMNLILRVEWSKPKVKE 274
> cel:F26A3.2 ncbp-2; Nuclear Cap-Binding Protein family member
(ncbp-2); K12883 nuclear cap-binding protein subunit 2
Length=154
Score = 56.2 bits (134), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/88 (32%), Positives = 46/88 (52%), Gaps = 0/88 (0%)
Query 235 ALKDLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFIT 294
++D E R T+ V NLS KE+ + ELFG G + RV + + KK GF F+
Sbjct 25 TVRDQETALRTSCTLYVGNLSYYTKEDQVYELFGRAGDVRRVIMGLDRFKKTPCGFCFVE 84
Query 295 YANRELAAQAIRKLNRHGYDNLLLNVEW 322
Y RE A A++ ++ D+ ++ +W
Sbjct 85 YYTREDAELALQNISNTRMDDRVIRADW 112
> xla:100380998 ncbp2; nuclear cap binding protein subunit 2,
20kDa; K12883 nuclear cap-binding protein subunit 2
Length=153
Score = 55.1 bits (131), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/85 (34%), Positives = 46/85 (54%), Gaps = 0/85 (0%)
Query 238 DLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYAN 297
D E+ ++ T+ V NLS EE + ELF G ++R+ + K KK + GF F+ Y
Sbjct 28 DQESLLKQSCTLYVGNLSFYTTEEQIHELFSKSGDVKRIVMGLDKVKKTACGFCFVEYYT 87
Query 298 RELAAQAIRKLNRHGYDNLLLNVEW 322
R A QA+R +N D+ ++ +W
Sbjct 88 RADAEQAMRFINGTRLDDRIVRTDW 112
> ath:AT2G44710 RNA recognition motif (RRM)-containing protein
Length=809
Score = 53.5 bits (127), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 46/69 (66%), Gaps = 0/69 (0%)
Query 240 EARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRE 299
E R+R+E I V +L + A EEDL ++FG++G++ V + K+ K SKG AF+ +A E
Sbjct 207 ERRKRKEFEIFVGSLDKGASEEDLKKVFGHVGEVTEVRILKNPQTKKSKGSAFLRFATVE 266
Query 300 LAAQAIRKL 308
A +A+++L
Sbjct 267 QAKRAVKEL 275
Score = 35.4 bits (80), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 36/82 (43%), Gaps = 4/82 (4%)
Query 248 TIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRK 307
TI + L EE + +L GKLE+V LA++ K F F+T+ E A +
Sbjct 392 TIFIDGLLPSWNEERVRDLLKPYGKLEKVELARNMPSARRKDFGFVTFDTHEAAVSCAKF 451
Query 308 LNR----HGYDNLLLNVEWARP 325
+N G D + +RP
Sbjct 452 INNSELGEGEDKAKVRARLSRP 473
> ath:AT4G34110 PAB2; PAB2 (POLY(A) BINDING 2); RNA binding /
translation initiation factor; K13126 polyadenylate-binding
protein
Length=629
Score = 53.5 bits (127), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 44/79 (55%), Gaps = 1/79 (1%)
Query 237 KDLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYA 296
+D A + + + V NL+E ++DL FG GK+ + K + K SKGF F+ +
Sbjct 205 RDSTANKTKFTNVYVKNLAESTTDDDLKNAFGEYGKITSAVVMKDGEGK-SKGFGFVNFE 263
Query 297 NRELAAQAIRKLNRHGYDN 315
N + AA+A+ LN H +D+
Sbjct 264 NADDAARAVESLNGHKFDD 282
Score = 44.7 bits (104), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 40/83 (48%), Gaps = 6/83 (7%)
Query 237 KDLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYA 296
+D RR I + NL E + L + F + G + +A + SKG+ F+ YA
Sbjct 114 RDPSVRRSGAGNIFIKNLDESIDHKALHDTFSSFGNIVSCKVAVDSSGQ-SKGYGFVQYA 172
Query 297 NRELAAQAIRKLNRHGYDNLLLN 319
N E A +AI KLN +LLN
Sbjct 173 NEESAQKAIEKLN-----GMLLN 190
> ath:AT2G21440 RNA recognition motif (RRM)-containing protein;
K14573 nucleolar protein 4
Length=1003
Score = 53.1 bits (126), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 0/77 (0%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
+ NL AK D+ +F +G + VF+ K+ + KGFAF+ + ++ AA AI+K N
Sbjct 335 IRNLPFQAKPSDIKVVFSAVGFVWDVFIPKNFETGLPKGFAFVKFTCKKDAANAIKKFNG 394
Query 311 HGYDNLLLNVEWARPNN 327
H + + V+WA P N
Sbjct 395 HMFGKRPIAVDWAVPKN 411
Score = 40.0 bits (92), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 41/86 (47%), Gaps = 2/86 (2%)
Query 247 ATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIR 306
AT+ V+ L L E F +G + R FL +K +GFAF+ +A +E +AI
Sbjct 20 ATVCVSGLPYSITNAQLEEAFSEVGPVRRCFLVTNKGSDEHRGFAFVKFALQEDVNRAIE 79
Query 307 KLNRHGYDNLLLNVEWA--RPNNRER 330
N + V+ A RP+ +ER
Sbjct 80 LKNGSTVGGRRITVKQAAHRPSLQER 105
Score = 32.7 bits (73), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 32/60 (53%), Gaps = 0/60 (0%)
Query 246 EATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAI 305
E T+ + NL D +E++ + F G++E + L HK K +G AF+ + + + AI
Sbjct 560 ERTLFIRNLPFDVTKEEVKQRFTVFGEVESLSLVLHKVTKRPEGTAFVKFKTADASVAAI 619
> mmu:68092 Ncbp2, 20kDa, 5930413E18Rik, AA536802, AI843301, C79367;
nuclear cap binding protein subunit 2; K12883 nuclear
cap-binding protein subunit 2
Length=156
Score = 50.8 bits (120), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 43/79 (54%), Gaps = 0/79 (0%)
Query 244 REEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQ 303
++ T+ V NLS EE + ELF G ++++ + K KK + GF F+ Y +R A
Sbjct 37 KKSCTLYVGNLSFYTTEEQIYELFSKSGDIKKIIMGLDKMKKTACGFCFVEYYSRADAEN 96
Query 304 AIRKLNRHGYDNLLLNVEW 322
A+R +N D+ ++ +W
Sbjct 97 AMRYINGTRLDDRIIRTDW 115
> ath:AT4G24770 RBP31; RBP31 (31-KDA RNA BINDING PROTEIN); RNA
binding / poly(U) binding
Length=329
Score = 50.8 bits (120), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 44/81 (54%), Gaps = 0/81 (0%)
Query 245 EEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQA 304
EEA + V NL+ D + LA LF G +E + +++ S+GF F+T ++ + A A
Sbjct 148 EEAKLFVGNLAYDVNSQALAMLFEQAGTVEIAEVIYNRETDQSRGFGFVTMSSVDEAETA 207
Query 305 IRKLNRHGYDNLLLNVEWARP 325
+ K NR+ + LL V A P
Sbjct 208 VEKFNRYDLNGRLLTVNKAAP 228
Score = 32.0 bits (71), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 38/82 (46%), Gaps = 2/82 (2%)
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
+ V NL D L +LF GK+ + ++ S+GF F+T ++ + +AI L
Sbjct 246 VYVGNLPWDVDNGRLEQLFSEHGKVVEARVVYDRETGRSRGFGFVTMSDVDELNEAISAL 305
Query 309 NRHGYDN--LLLNVEWARPNNR 328
+ + + +NV RP R
Sbjct 306 DGQNLEGRAIRVNVAEERPPRR 327
> dre:192325 ncbp2, CHUNP6917, zgc:92592; nuclear cap binding
protein subunit 2; K12883 nuclear cap-binding protein subunit
2
Length=155
Score = 50.8 bits (120), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 42/79 (53%), Gaps = 0/79 (0%)
Query 244 REEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQ 303
++ AT+ V NLS EE + LF G ++R+ + K KK + GF F+ Y R A
Sbjct 36 KQSATLYVGNLSFYTTEEQVHVLFAKCGDVKRIIIGLDKIKKTACGFCFVEYYTRADAEN 95
Query 304 AIRKLNRHGYDNLLLNVEW 322
A+R +N D+ ++ +W
Sbjct 96 AMRFVNGTRLDDRIIRTDW 114
> ath:AT3G10400 RNA recognition motif (RRM)-containing protein;
K13154 U11/U12 small nuclear ribonucleoprotein 31 kDa protein
Length=261
Score = 50.1 bits (118), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 44/83 (53%), Gaps = 0/83 (0%)
Query 246 EATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAI 305
++T+ V+NL D+ LF GK+ RV + K + + S+G AF+ Y +RE AA+A
Sbjct 56 KSTLYVSNLDFSLTNSDIHTLFSTFGKVARVTVLKDRHTRQSRGVAFVLYVSREDAAKAA 115
Query 306 RKLNRHGYDNLLLNVEWARPNNR 328
R ++ + L V A N R
Sbjct 116 RSMDAKILNGRKLTVSIAADNGR 138
> ath:AT3G52660 RNA recognition motif (RRM)-containing protein
Length=471
Score = 49.3 bits (116), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 38/61 (62%), Gaps = 0/61 (0%)
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
+ + + DA E DL G+IG++ V + + KD KG+AF+T+ +++LAA+AI L
Sbjct 94 VYLGGIPTDATEGDLKGFCGSIGEVTEVRIMREKDSGDGKGYAFVTFRSKDLAAEAIDTL 153
Query 309 N 309
N
Sbjct 154 N 154
Score = 43.1 bits (100), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 38/78 (48%), Gaps = 0/78 (0%)
Query 248 TIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRK 307
+ + NL D +E L LF + GK+ +V + K K + F+ YA R +A++
Sbjct 269 ALYIKNLPRDITQERLKALFEHHGKILKVVIPPAKPGKEDSRYGFVHYAERTSVMRALKN 328
Query 308 LNRHGYDNLLLNVEWARP 325
R+ D +L+ A+P
Sbjct 329 TERYEIDGHMLDCTLAKP 346
> hsa:1994 ELAVL1, ELAV1, HUR, Hua, MelG; ELAV (embryonic lethal,
abnormal vision, Drosophila)-like 1 (Hu antigen R); K13088
ELAV-like protein 1
Length=326
Score = 49.3 bits (116), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 34/107 (31%), Positives = 50/107 (46%), Gaps = 8/107 (7%)
Query 216 SRLGVPGAKGLSRLRGGGDALKDLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLER 275
S +GV GLS + G+A I + NL +DA E L ++FG G +
Sbjct 221 SPMGVDHMSGLSGVNVPGNASSGW--------CIFIYNLGQDADEGILWQMFGPFGAVTN 272
Query 276 VFLAKHKDKKASKGFAFITYANRELAAQAIRKLNRHGYDNLLLNVEW 322
V + + + KGF F+T N E AA AI LN + + +L V +
Sbjct 273 VKVIRDFNTNKCKGFGFVTMTNYEEAAMAIASLNGYRLGDKILQVSF 319
Score = 32.3 bits (72), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 16/67 (23%), Positives = 33/67 (49%), Gaps = 0/67 (0%)
Query 245 EEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQA 304
++A + ++ L ++D+ ++F G++ + + S+G AFI + R A +A
Sbjct 104 KDANLYISGLPRTMTQKDVEDMFSRFGRIINSRVLVDQTTGLSRGVAFIRFDKRSEAEEA 163
Query 305 IRKLNRH 311
I N H
Sbjct 164 ITSFNGH 170
> mmu:19652 Rbm3, 2600016C11Rik, MGC118410; RNA binding motif
protein 3; K13186 RNA-binding protein 3
Length=154
Score = 48.5 bits (114), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 43/79 (54%), Gaps = 0/79 (0%)
Query 245 EEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQA 304
EE + V L+ + E+ L + F + G + V + K ++ + S+GF FIT+ N E A+ A
Sbjct 4 EEGKLFVGGLNFNTDEQALEDHFSSFGPISEVVVVKDRETQRSRGFGFITFTNPEHASDA 63
Query 305 IRKLNRHGYDNLLLNVEWA 323
+R +N D + V+ A
Sbjct 64 MRAMNGESLDGRQIRVDHA 82
> mmu:100043257 Gm15453, OTTMUSG00000022091; predicted gene 15453
Length=154
Score = 48.5 bits (114), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 43/79 (54%), Gaps = 0/79 (0%)
Query 245 EEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQA 304
EE + V L+ + E+ L + F + G + V + K ++ + S+GF FIT+ N E A+ A
Sbjct 4 EEGKLFVGGLNFNTDEQALEDHFSSFGPISEVVVVKDRETQRSRGFGFITFTNPEHASDA 63
Query 305 IRKLNRHGYDNLLLNVEWA 323
+R +N D + V+ A
Sbjct 64 MRAMNGESLDGRQIRVDHA 82
> mmu:15572 Elavl4, Elav, Hud, PNEM; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 4 (Hu antigen D); K13208 ELAV
like protein 2/3/4
Length=380
Score = 48.5 bits (114), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
I V NLS D+ E L +LFG G + V + + + KGF F+T N + AA AI L
Sbjct 299 IFVYNLSPDSDESVLWQLFGPFGAVNNVKVIRDFNTNKCKGFGFVTMTNYDEAAMAIASL 358
Query 309 NRHGYDNLLLNVEW 322
N + + +L V +
Sbjct 359 NGYRLGDRVLQVSF 372
Score = 45.8 bits (107), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 41/77 (53%), Gaps = 0/77 (0%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
V L ++ +E+ LFG+IG++E L + K S G+ F+ Y + + A +AI LN
Sbjct 50 VNYLPQNMTQEEFRSLFGSIGEIESCKLVRDKITGQSLGYGFVNYIDPKDAEKAINTLNG 109
Query 311 HGYDNLLLNVEWARPNN 327
+ V +ARP++
Sbjct 110 LRLQTKTIKVSYARPSS 126
> mmu:15568 Elavl1, 2410055N02Rik, HUR, Hua, W91709; ELAV (embryonic
lethal, abnormal vision, Drosophila)-like 1 (Hu antigen
R); K13088 ELAV-like protein 1
Length=326
Score = 48.5 bits (114), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 33/107 (30%), Positives = 50/107 (46%), Gaps = 8/107 (7%)
Query 216 SRLGVPGAKGLSRLRGGGDALKDLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLER 275
S +GV G+S + G+A I + NL +DA E L ++FG G +
Sbjct 221 SPMGVDHMSGISGVNVPGNASSGW--------CIFIYNLGQDADEGILWQMFGPFGAVTN 272
Query 276 VFLAKHKDKKASKGFAFITYANRELAAQAIRKLNRHGYDNLLLNVEW 322
V + + + KGF F+T N E AA AI LN + + +L V +
Sbjct 273 VKVIRDFNTNKCKGFGFVTMTNYEEAAMAIASLNGYRLGDKILQVSF 319
Score = 42.7 bits (99), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 41/77 (53%), Gaps = 0/77 (0%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
V L ++ +E+L LF +IG++E L + K S G+ F+ Y + A +AI LN
Sbjct 24 VNYLPQNMTQEELRSLFSSIGEVESAKLIRDKVAGHSLGYGFVNYVTAKDAERAISTLNG 83
Query 311 HGYDNLLLNVEWARPNN 327
+ + V +ARP++
Sbjct 84 LRLQSKTIKVSYARPSS 100
Score = 32.3 bits (72), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 16/67 (23%), Positives = 33/67 (49%), Gaps = 0/67 (0%)
Query 245 EEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQA 304
++A + ++ L ++D+ ++F G++ + + S+G AFI + R A +A
Sbjct 104 KDANLYISGLPRTMTQKDVEDMFSRFGRIINSRVLVDQTTGLSRGVAFIRFDKRSEAEEA 163
Query 305 IRKLNRH 311
I N H
Sbjct 164 ITSFNGH 170
> ath:AT4G19610 RNA binding / nucleic acid binding / nucleotide
binding; K14787 multiple RNA-binding domain-containing protein
1
Length=816
Score = 48.5 bits (114), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 39/70 (55%), Gaps = 0/70 (0%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
V NL A EE+L E F GK+ V L K+ K S+G A+I Y E AA+A+ +L+
Sbjct 298 VRNLPYTATEEELMEHFSTFGKISEVHLVLDKETKRSRGIAYILYLIPECAARAMEELDN 357
Query 311 HGYDNLLLNV 320
+ LL++
Sbjct 358 SSFQGRLLHI 367
Score = 34.7 bits (78), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 21/79 (26%), Positives = 42/79 (53%), Gaps = 1/79 (1%)
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
+ V N++ +A + +L +LF G+++ + L K K+ G+AF+ + ++ A A + L
Sbjct 704 LHVKNIAFEATKRELRQLFSPFGQIKSMRLPK-KNIGQYAGYAFVEFVTKQEALNAKKAL 762
Query 309 NRHGYDNLLLNVEWARPNN 327
+ L +EWA +N
Sbjct 763 ASTHFYGRHLVLEWANDDN 781
Score = 34.3 bits (77), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 37/75 (49%), Gaps = 1/75 (1%)
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
I V NL + KE+ L + F G++ L + D K S+ F FI + + + A QAI+
Sbjct 4 ICVKNLPKHVKEDQLRDHFSQKGEITDAKLMRSNDGK-SRQFGFIGFRSAQEAQQAIKYF 62
Query 309 NRHGYDNLLLNVEWA 323
N L+ VE A
Sbjct 63 NNTYLGTSLIIVEIA 77
> dre:30737 elavl4, elrd, wu:fc24h01, zHuD; ELAV (embryonic lethal,
abnormal vision, Drosophila)-like 4 (Hu antigen D); K13208
ELAV like protein 2/3/4
Length=367
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
I V NLS D+ E L +LFG G + V + + + KGF F+T N + AA AI L
Sbjct 286 IFVYNLSPDSDESVLWQLFGPFGAVNNVKVIRDFNTNKCKGFGFVTMTNYDEAAMAIASL 345
Query 309 NRHGYDNLLLNVEW 322
N + + +L V +
Sbjct 346 NGYRLGDRVLQVSF 359
Score = 45.4 bits (106), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 41/77 (53%), Gaps = 0/77 (0%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
V L ++ +E+ LFG+IG++E L + K S G+ F+ Y + + A +AI LN
Sbjct 48 VNYLPQNMTQEEFRSLFGSIGEIESCKLVRDKITGQSLGYGFVNYIDPKDAEKAINTLNG 107
Query 311 HGYDNLLLNVEWARPNN 327
+ V +ARP++
Sbjct 108 LRLQTKTIKVSYARPSS 124
> dre:30065 hug, fb72d01, fk65g02, wu:fb72d01, wu:fk65g02, zHuG;
HuG; K13208 ELAV like protein 2/3/4
Length=322
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
I V NL +DA E L ++FG G + V + + + KGF F+T N E AA AI L
Sbjct 242 IFVYNLGQDADEGILWQMFGPFGAVTNVKVIRDFNTSKCKGFGFVTMTNYEEAAMAISSL 301
Query 309 NRHGYDNLLLNVEW 322
N + + +L V +
Sbjct 302 NGYRLGDKVLQVSF 315
Score = 45.1 bits (105), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 41/77 (53%), Gaps = 0/77 (0%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
+ L ++ +E+L LF +IG++E L + K S G+ F+ Y N A +AI LN
Sbjct 22 INYLPQNMSQEELRSLFSSIGEVESAKLIRDKMAGHSLGYGFVNYVNPSDAERAINTLNG 81
Query 311 HGYDNLLLNVEWARPNN 327
+ + V +ARP++
Sbjct 82 LRLQSKTIKVSYARPSS 98
Score = 31.6 bits (70), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 20/84 (23%), Positives = 40/84 (47%), Gaps = 8/84 (9%)
Query 226 LSRLRGGGDALKDLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKK 285
+S R D +KD A + ++ L + ++++ ++F G++ + +
Sbjct 91 VSYARPSSDGIKD--------ANLYISGLPKTMTQKNVEDMFTQYGRIINSRILVDQASG 142
Query 286 ASKGFAFITYANRELAAQAIRKLN 309
S+G AFI + R A +AI+ LN
Sbjct 143 LSRGVAFIRFDKRSEAEEAIKDLN 166
> xla:386602 elavl4, HuD, elrD, elrD1, elrD2; ELAV (embryonic
lethal, abnormal vision)-like 4 (Hu antigen D); K13208 ELAV
like protein 2/3/4
Length=400
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
I V NLS D+ E L +LFG G + V + + + KGF F+T N + AA AI L
Sbjct 319 IFVYNLSPDSDESVLWQLFGPFGAVNNVKVIRDFNTNKCKGFGFVTMTNYDEAAMAIASL 378
Query 309 NRHGYDNLLLNVEW 322
N + + +L V +
Sbjct 379 NGYRLGDRVLQVSF 392
> hsa:1996 ELAVL4, HUD, PNEM; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 4 (Hu antigen D); K13208 ELAV like
protein 2/3/4
Length=366
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
I V NLS D+ E L +LFG G + V + + + KGF F+T N + AA AI L
Sbjct 285 IFVYNLSPDSDESVLWQLFGPFGAVNNVKVIRDFNTNKCKGFGFVTMTNYDEAAMAIASL 344
Query 309 NRHGYDNLLLNVEW 322
N + + +L V +
Sbjct 345 NGYRLGDRVLQVSF 358
Score = 45.4 bits (106), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 41/77 (53%), Gaps = 0/77 (0%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
V L ++ +E+ LFG+IG++E L + K S G+ F+ Y + + A +AI LN
Sbjct 50 VNYLPQNMTQEEFRSLFGSIGEIESCKLVRDKITGQSLGYGFVNYIDPKDAEKAINTLNG 109
Query 311 HGYDNLLLNVEWARPNN 327
+ V +ARP++
Sbjct 110 LRLQTKTIKVSYARPSS 126
> xla:446310 zcrb1, MGC82154; zinc finger CCHC-type and RNA binding
motif 1; K13154 U11/U12 small nuclear ribonucleoprotein
31 kDa protein
Length=218
Score = 48.1 bits (113), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 42/83 (50%), Gaps = 0/83 (0%)
Query 246 EATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAI 305
++T+ V+NL DL +F GK+ +V + K KD + SKG +F+ + ++E A +
Sbjct 9 KSTVYVSNLPFSLTNNDLHRIFSKYGKVVKVTILKDKDSRKSKGVSFVLFLDKESAQNCV 68
Query 306 RKLNRHGYDNLLLNVEWARPNNR 328
R LN + A+ N R
Sbjct 69 RGLNNKQLFGRAIKASIAKDNGR 91
> hsa:5935 RBM3, IS1-RNPL, RNPL; RNA binding motif (RNP1, RRM)
protein 3; K13186 RNA-binding protein 3
Length=157
Score = 47.8 bits (112), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 43/79 (54%), Gaps = 0/79 (0%)
Query 245 EEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQA 304
EE + V L+ + E+ L + F + G + V + K ++ + S+GF FIT+ N E A+ A
Sbjct 4 EEGKLFVGGLNFNTDEQALEDHFSSFGPISEVVVVKDRETQRSRGFGFITFTNPEHASVA 63
Query 305 IRKLNRHGYDNLLLNVEWA 323
+R +N D + V+ A
Sbjct 64 MRAMNGESLDGRQIRVDHA 82
> hsa:1153 CIRBP, CIRP; cold inducible RNA binding protein; K13195
cold-inducible RNA-binding protein
Length=172
Score = 47.8 bits (112), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 26/88 (29%), Positives = 46/88 (52%), Gaps = 2/88 (2%)
Query 245 EEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQA 304
+E + V LS D E+ L ++F G++ V + K ++ + S+GF F+T+ N + A A
Sbjct 4 DEGKLFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDA 63
Query 305 IRKLNRHGYDNLLLNVEWA--RPNNRER 330
+ +N D + V+ A +NR R
Sbjct 64 MMAMNGKSVDGRQIRVDQAGKSSDNRSR 91
> ath:AT1G49600 ATRBP47A (Arabidopsis thaliana RNA-binding protein
47a); RNA binding
Length=445
Score = 47.8 bits (112), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 49/104 (47%), Gaps = 9/104 (8%)
Query 226 LSRLRGGGDALKDLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKK 285
L+ GG ++ D E+ +TI V L D EEDL + F + G++ V +
Sbjct 309 LAGGHGGNGSMSDGESNN---STIFVGGLDADVTEEDLMQPFSDFGEVVSVKI------P 359
Query 286 ASKGFAFITYANRELAAQAIRKLNRHGYDNLLLNVEWARPNNRE 329
KG F+ +ANR+ A +AI LN + + W R N++
Sbjct 360 VGKGCGFVQFANRQSAEEAIGNLNGTVIGKNTVRLSWGRSPNKQ 403
> dre:321860 rbmx, fc66e11, wu:fb37c07, wu:fc66e11; RNA binding
motif protein, X-linked; K12885 heterogeneous nuclear ribonucleoprotein
G
Length=379
Score = 47.4 bits (111), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 37/73 (50%), Gaps = 0/73 (0%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
+ L+ + E+ L F G++ V L K ++ S+GFAF+TY N A A R++N
Sbjct 12 IGGLNTETSEKVLEAYFSKFGRISEVLLMKDRETNKSRGFAFVTYENPGDAKDAAREMNG 71
Query 311 HGYDNLLLNVEWA 323
D + VE A
Sbjct 72 KPLDGKPIKVEQA 84
> hsa:1993 ELAVL2, HEL-N1, HELN1, HUB; ELAV (embryonic lethal,
abnormal vision, Drosophila)-like 2 (Hu antigen B); K13208
ELAV like protein 2/3/4
Length=346
Score = 47.4 bits (111), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 42/77 (54%), Gaps = 0/77 (0%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
V L ++ +E+L LFG+IG++E L + K S G+ F+ Y + + A +AI LN
Sbjct 43 VNYLPQNMTQEELKSLFGSIGEIESCKLVRDKITGQSLGYGFVNYIDPKDAEKAINTLNG 102
Query 311 HGYDNLLLNVEWARPNN 327
+ V +ARP++
Sbjct 103 LRLQTKTIKVSYARPSS 119
Score = 47.4 bits (111), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
I V NL+ DA E L ++FG G + V + + + KGF F+T N + AA AI L
Sbjct 265 IFVYNLAPDADESILWQMFGPFGAVTNVKVIRDFNTNKCKGFGFVTMTNYDEAAMAIASL 324
Query 309 NRHGYDNLLLNVEW 322
N + + +L V +
Sbjct 325 NGYRLGDRVLQVSF 338
> mmu:15569 Elavl2, Hub, mel-N1; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 2 (Hu antigen B); K13208 ELAV like
protein 2/3/4
Length=359
Score = 47.4 bits (111), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 42/77 (54%), Gaps = 0/77 (0%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
V L ++ +E+L LFG+IG++E L + K S G+ F+ Y + + A +AI LN
Sbjct 43 VNYLPQNMTQEELKSLFGSIGEIESCKLVRDKITGQSLGYGFVNYIDPKDAEKAINTLNG 102
Query 311 HGYDNLLLNVEWARPNN 327
+ V +ARP++
Sbjct 103 LRLQTKTIKVSYARPSS 119
Score = 47.4 bits (111), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
I V NL+ DA E L ++FG G + V + + + KGF F+T N + AA AI L
Sbjct 278 IFVYNLAPDADESILWQMFGPFGAVTNVKVIRDFNTNKCKGFGFVTMTNYDEAAMAIASL 337
Query 309 NRHGYDNLLLNVEW 322
N + + +L V +
Sbjct 338 NGYRLGDRVLQVSF 351
> ath:AT2G35410 33 kDa ribonucleoprotein, chloroplast, putative
/ RNA-binding protein cp33, putative
Length=308
Score = 47.4 bits (111), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 41/74 (55%), Gaps = 1/74 (1%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
V NL D++ELFG G + V + + KD K ++GFAF+T A+ E A AI K +
Sbjct 99 VFNLPWSMSVNDISELFGQCGTVNNVEIIRQKDGK-NRGFAFVTMASGEEAQAAIDKFDT 157
Query 311 HGYDNLLLNVEWAR 324
+++V +AR
Sbjct 158 FQVSGRIISVSFAR 171
> xla:397952 elavl2-b, elavl2, elrB, hel-n1, heln1, hub, xel-1;
ELAV (embryonic lethal, abnormal vision)-like 2 (Hu antigen
B); K13208 ELAV like protein 2/3/4
Length=389
Score = 47.0 bits (110), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
I V NL+ DA E L ++FG G + V + + + KGF F+T N + AA AI L
Sbjct 308 IFVYNLAPDADESILWQMFGPFGAVTNVKVIRDFNTNKCKGFGFVTMTNYDEAAMAIASL 367
Query 309 NRHGYDNLLLNVEW 322
N + + +L V +
Sbjct 368 NGYRLGDRVLQVSF 381
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 43/78 (55%), Gaps = 1/78 (1%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKA-SKGFAFITYANRELAAQAIRKLN 309
V L ++ +E+L LFG+IG++E L + K + S G+ F+ Y + + A +AI +N
Sbjct 70 VNYLPQNMTQEELKSLFGSIGEIESCKLVRDKITEGQSLGYGFVNYIDPKDAEKAINTVN 129
Query 310 RHGYDNLLLNVEWARPNN 327
+ V +ARP++
Sbjct 130 GLRLQTKTIKVSYARPSS 147
> xla:394342 elavl2-a, Xel-1, el-1, elavl2, elrB, hel-n1, heln1,
hub, p45; ELAV (embryonic lethal, abnormal vision)-like 2
(Hu antigen B); K13208 ELAV like protein 2/3/4
Length=389
Score = 47.0 bits (110), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 39/74 (52%), Gaps = 0/74 (0%)
Query 249 IRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKL 308
I V NL+ DA E L ++FG G + V + + + KGF F+T N + AA AI L
Sbjct 308 IFVYNLAPDADESILWQMFGPFGAVTNVKVIRDFNTNKCKGFGFVTMTNYDEAAMAIASL 367
Query 309 NRHGYDNLLLNVEW 322
N + + +L V +
Sbjct 368 NGYRLGDRVLQVSF 381
Score = 43.5 bits (101), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 43/78 (55%), Gaps = 1/78 (1%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKA-SKGFAFITYANRELAAQAIRKLN 309
V L ++ +E+L LFG+IG++E L + K + S G+ F+ Y + + A +AI LN
Sbjct 70 VNYLPQNMTQEELKSLFGSIGEIESCKLVRDKITEGQSLGYGFVNYIDPKDAEKAINTLN 129
Query 310 RHGYDNLLLNVEWARPNN 327
+ V +ARP++
Sbjct 130 GLRLQTKTIKVSYARPSS 147
> hsa:9904 RBM19, DKFZp586F1023, KIAA0682; RNA binding motif protein
19; K14787 multiple RNA-binding domain-containing protein
1
Length=960
Score = 47.0 bits (110), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 41/77 (53%), Gaps = 3/77 (3%)
Query 247 ATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKA---SKGFAFITYANRELAAQ 303
T+ + NL+ D EE L E+F +G ++ ++K K+K S GF F+ Y E A +
Sbjct 730 CTLFIKNLNFDTTEEKLKEVFSKVGTVKSCSISKKKNKAGVLLSMGFGFVEYRKPEQAQK 789
Query 304 AIRKLNRHGYDNLLLNV 320
A+++L H D L V
Sbjct 790 ALKQLQGHVVDGHKLEV 806
Score = 40.8 bits (94), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 37/76 (48%), Gaps = 0/76 (0%)
Query 245 EEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQA 304
E + V NL + EEDL +LF G L + K KGFAFIT+ E A +A
Sbjct 400 ESGRLFVRNLPYTSTEEDLEKLFSKYGPLSELHYPIDSLTKKPKGFAFITFMFPEHAVKA 459
Query 305 IRKLNRHGYDNLLLNV 320
+++ + +L+V
Sbjct 460 YSEVDGQVFQGRMLHV 475
Score = 36.2 bits (82), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 34/74 (45%), Gaps = 1/74 (1%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
V NL KEE +LF G L L KD K K F FI + + E A +A + N+
Sbjct 6 VKNLPNGMKEERFRQLFAAFGTLTDCSLKFTKDGKFRK-FGFIGFKSEEEAQKAQKHFNK 64
Query 311 HGYDNLLLNVEWAR 324
D + VE+ +
Sbjct 65 SFIDTSRITVEFCK 78
Score = 35.4 bits (80), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 43/85 (50%), Gaps = 4/85 (4%)
Query 242 RRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKAS-KGFAFITYANREL 300
R++ + I V N+ A ++ ELF G+L+ V L K + +GF F+ + ++
Sbjct 827 RKQTTSKILVRNIPFQAHSREIRELFSTFGELKTVRLPKKMTGTGTHRGFGFVDFLTKQD 886
Query 301 AAQAIRKL--NRHGYDNLLLNVEWA 323
A +A L + H Y L+ +EWA
Sbjct 887 AKRAFNALCHSTHLYGRRLV-LEWA 910
> sce:YPL178W CBC2, CBP20, MUD13, SAE1; Cbc2p; K12883 nuclear
cap-binding protein subunit 2
Length=208
Score = 47.0 bits (110), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 50/105 (47%), Gaps = 1/105 (0%)
Query 217 RLGVPGAKGLSRLRGGGDALKDLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERV 276
RL P L + R + L++L + +TI V NLS EE + ELF G ++R+
Sbjct 17 RLDTPSRYLLRKARRNPNGLQELRESMKS-STIYVGNLSFYTSEEQIYELFSKCGTIKRI 75
Query 277 FLAKHKDKKASKGFAFITYANRELAAQAIRKLNRHGYDNLLLNVE 321
+ + K GF FI Y+ + A A++ L+ D + ++
Sbjct 76 IMGLDRFKFTPCGFCFIIYSCPDEALNALKYLSDTKLDEKTITID 120
> ath:AT4G20030 RNA recognition motif (RRM)-containing protein
Length=152
Score = 47.0 bits (110), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 45/83 (54%), Gaps = 0/83 (0%)
Query 247 ATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIR 306
+ I V NL E+ L F G++ V L K + K SKG+AFI + +++ A AI
Sbjct 40 SKIMVRNLPFSTSEDFLKREFSAFGEIAEVKLIKDEAMKRSKGYAFIQFTSQDDAFLAIE 99
Query 307 KLNRHGYDNLLLNVEWARPNNRE 329
++R Y+ ++ ++ A+P R+
Sbjct 100 TMDRRMYNGRMIYIDIAKPGKRD 122
> xla:379484 cirbp-b, MGC64461, Xcirp, cirbp, cirp, cirp2, xcirp2;
cold inducible RNA binding protein; K13195 cold-inducible
RNA-binding protein
Length=166
Score = 47.0 bits (110), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 42/79 (53%), Gaps = 0/79 (0%)
Query 245 EEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQA 304
+E + + L+ D EE L ++F G++ V + K ++ K S+GF F+T+ N + A A
Sbjct 3 DEGKLFIGGLNFDTNEESLEQVFSKYGQISEVVVVKDRETKRSRGFGFVTFENPDDAKDA 62
Query 305 IRKLNRHGYDNLLLNVEWA 323
+ +N D + V+ A
Sbjct 63 MMAMNGKAVDGRQIRVDQA 81
> dre:431719 elavl2, zgc:91918; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 2 (Hu antigen B); K13208 ELAV like
protein 2/3/4
Length=360
Score = 47.0 bits (110), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 41/77 (53%), Gaps = 0/77 (0%)
Query 251 VTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRKLNR 310
V L ++ +E+L LFG+IG++E L + K S G+ F+ Y + A +AI LN
Sbjct 43 VNYLPQNMTQEELKSLFGSIGEIESCKLVRDKITGQSLGYGFVNYMEPKDAEKAINTLNG 102
Query 311 HGYDNLLLNVEWARPNN 327
+ V +ARP++
Sbjct 103 LRLQTKTIKVSYARPSS 119
Score = 47.0 bits (110), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 42/85 (49%), Gaps = 0/85 (0%)
Query 238 DLEARRREEATIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYAN 297
+L A I V NL+ DA E L ++FG G + V + + + KGF F+T N
Sbjct 268 NLPAHAGTGWCIFVYNLAPDADENVLWQMFGPFGAVTNVKVIRDFNTNKCKGFGFVTMTN 327
Query 298 RELAAQAIRKLNRHGYDNLLLNVEW 322
+ AA AI LN + + +L V +
Sbjct 328 YDEAAVAIASLNGYRLGDRVLQVSF 352
> xla:399292 elavl1-a, Elavl1, MGC132161, Xel-1, elav1, elrA,
hua, hur, melg, p36; ELAV (embryonic lethal, abnormal vision)-like
1 (Hu antigen R); K13208 ELAV like protein 2/3/4
Length=337
Score = 47.0 bits (110), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 38/75 (50%), Gaps = 0/75 (0%)
Query 248 TIRVTNLSEDAKEEDLAELFGNIGKLERVFLAKHKDKKASKGFAFITYANRELAAQAIRK 307
I V NL +DA E L ++FG G + V + + + KGF F+T N E AA AI
Sbjct 256 CIFVYNLGQDADEGILWQMFGPFGAVTNVKVIRDFNTNKCKGFGFVTMTNYEEAAMAIAS 315
Query 308 LNRHGYDNLLLNVEW 322
LN + + L V +
Sbjct 316 LNGYRLGDKTLQVSF 330
Lambda K H
0.317 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 13775884780
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40