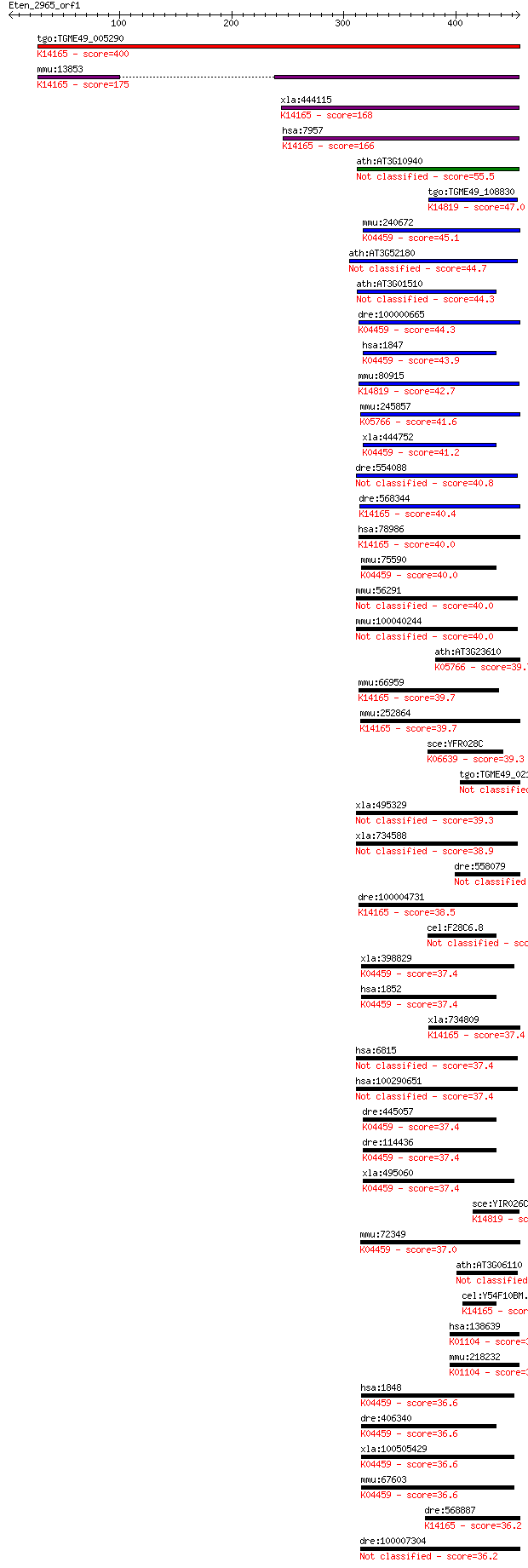
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2965_orf1
Length=456
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_005290 dual-specificity phosphatase laforin, putati... 400 7e-111
mmu:13853 Epm2a; epilepsy, progressive myoclonic epilepsy, typ... 175 3e-43
xla:444115 epm2a, MGC80469; epilepsy, progressive myoclonus ty... 168 4e-41
hsa:7957 EPM2A, EPM2, MELF; epilepsy, progressive myoclonus ty... 166 2e-40
ath:AT3G10940 protein phosphatase-related 55.5 5e-07
tgo:TGME49_108830 dual specificity phosphatase, catalytic doma... 47.0 2e-04
mmu:240672 Dusp5, Gm337; dual specificity phosphatase 5; K0445... 45.1 6e-04
ath:AT3G52180 SEX4; SEX4 (STARCH-EXCESS 4); polysaccharide bin... 44.7 8e-04
ath:AT3G01510 5'-AMP-activated protein kinase beta-1 subunit-r... 44.3 0.001
dre:100000665 dual specificity phosphatase 3-like; K04459 dual... 44.3 0.001
hsa:1847 DUSP5, DUSP, HVH3; dual specificity phosphatase 5 (EC... 43.9 0.001
mmu:80915 Dusp12, 1190004O14Rik, AA027408, AW049275, ESTM36, L... 42.7 0.003
mmu:245857 Ssh3, BC028922, SSH-3, mSSH-3L; slingshot homolog 3... 41.6 0.006
xla:444752 dusp5, MGC84792; dual specificity phosphatase 5; K0... 41.2 0.009
dre:554088 styx, im:6908851, si:dkey-119m7.5, zgc:110621; seri... 40.8 0.011
dre:568344 dusp3, si:bz18k17.3, si:rp71-18k17.3; dual specific... 40.4 0.016
hsa:78986 DUSP26, DUSP24, LDP-4, MGC1136, MGC2627, MKP8, NATA1... 40.0 0.017
mmu:75590 Dusp9, Dusp4, Mpk4, Pyst3; dual specificity phosphat... 40.0 0.019
mmu:56291 Styx, 0610039A20Rik, MGC106795, hStyxb; serine/threo... 40.0 0.020
mmu:100040244 Gm14698, OTTMUSG00000017743; predicted gene 14698 40.0
ath:AT3G23610 dual specificity protein phosphatase (DsPTP1); K... 39.7 0.023
mmu:66959 Dusp26, 2310043K02Rik, Skrp3; dual specificity phosp... 39.7 0.023
mmu:252864 Dusp15, AI851682, LMW-DSP10, T-DSP10; dual specific... 39.7 0.025
sce:YFR028C CDC14, OAF3; Cdc14p (EC:3.1.3.48); K06639 cell div... 39.3 0.030
tgo:TGME49_021500 dual specificity protein phosphatase, cataly... 39.3 0.034
xla:495329 styx-b; serine/threonine/tyrosine interacting protein 39.3 0.036
xla:734588 styx-a, MGC115529, styx; serine/threonine/tyrosine ... 38.9 0.038
dre:558079 dual specificity phosphatase 8-like 38.9 0.045
dre:100004731 novel protein similar to vertebrate dual specifi... 38.5 0.055
cel:F28C6.8 hypothetical protein 38.5 0.056
xla:398829 dusp6, MGC68682, mkp3, pyst1; dual specificity phos... 37.4 0.12
hsa:1852 DUSP9, MKP-4, MKP4; dual specificity phosphatase 9 (E... 37.4 0.12
xla:734809 dusp18, MGC131167; dual specificity phosphatase 18;... 37.4 0.12
hsa:6815 STYX, FLJ42934; serine/threonine/tyrosine interacting... 37.4 0.12
hsa:100290651 serine/threonine/tyrosine-interacting protein-like 37.4 0.12
dre:445057 dusp2, wu:fj40g04, zgc:91929; dual specificity phos... 37.4 0.13
dre:114436 dusp5, wu:fc03b11, wu:fc86e02; dual specificity pho... 37.4 0.13
xla:495060 hypothetical LOC495060; K04459 dual specificity pho... 37.4 0.13
sce:YIR026C YVH1; Protein phosphatase involved in vegetative g... 37.4 0.14
mmu:72349 Dusp3, 2210015O03Rik, 5031436O03Rik, T-DSP11, VHR; d... 37.0 0.15
ath:AT3G06110 MKP2; MKP2 (MAPK PHOSPHATASE 2); MAP kinase phos... 37.0 0.16
cel:Y54F10BM.13 hypothetical protein; K14165 dual specificity ... 37.0 0.17
hsa:138639 PTPDC1, FLJ42922, PTP9Q22; protein tyrosine phospha... 37.0 0.18
mmu:218232 Ptpdc1, AI843923, AW456874, Naa-1; protein tyrosine... 37.0 0.18
hsa:1848 DUSP6, MKP3, PYST1; dual specificity phosphatase 6 (E... 36.6 0.20
dre:406340 dusp1, cb269, id:ibd2586, sb:cb269, wu:fd19g02, wu:... 36.6 0.21
xla:100505429 hypothetical protein LOC100505429; K04459 dual s... 36.6 0.21
mmu:67603 Dusp6, 1300019I03Rik, MGC98540, MKP-3, MKP3, PYST1; ... 36.6 0.21
dre:568887 zgc:172281 (EC:3.1.3.16); K14165 dual specificity p... 36.2 0.29
dre:100007304 dual specificity phosphatase 10-like 36.2 0.31
> tgo:TGME49_005290 dual-specificity phosphatase laforin, putative
(EC:3.1.3.16); K14165 dual specificity phosphatase [EC:3.1.3.16
3.1.3.48]
Length=523
Score = 400 bits (1027), Expect = 7e-111, Method: Compositional matrix adjust.
Identities = 232/489 (47%), Positives = 297/489 (60%), Gaps = 60/489 (12%)
Query 27 MRLRFSCVAFVPPGSQLGVSGGPDFLGRWDPCKCLPLLPHSSPSEGGIEPSLWFADVEVN 86
MR+RFS AFVPP +QLGV G FLG W C+PL+P+S+P G+EPSLWF D++++
Sbjct 1 MRVRFSVTAFVPPNAQLGVVGSAPFLGEWKLEHCVPLMPYSAPHPQGLEPSLWFRDIDLD 60
Query 87 EKS----LERCGKSAAGAAGCSS--HRCNGTEGAVDTF---------------------- 118
S +AA +A CS+ RC+ + + F
Sbjct 61 PASCPNDTNDVHSTAAVSASCSADRRRCDLSRQSPHCFPSARSASPYMVEVSRGDCPFAV 120
Query 119 -----SDKLIAAMTWTDSA--SAGTQR---SGFLPDTLSSTNDEASSIAA------AAYC 162
S +++ + +S SAG QR S + SS S++AA AA
Sbjct 121 ELMSTSQHVLSEFSKHNSPVYSAGEQRFRRSLPSSASFSSCTSSGSAVAATKCVWDAAAY 180
Query 163 RVDHRSPRIVAILEQHPLRRVQFEYKFILW---NSQKPVAPYVPDTPGESF--------- 210
RV R+P + A+LE HPLR FEYKF+LW N + V PYVP T GE
Sbjct 181 RVATRAPEVQAVLESHPLRHCAFEYKFVLWYPPNGEIAV-PYVPTTAGEEVPITYAGDSE 239
Query 211 ---QRSAYGSPSCTWRTWLFPPAPAQHAEPPSNSLHVIWEGSGPGTNRKFFFDPMDIVVD 267
+ S G+ +WR WLFPP+P++ PP S ++WEG GP +NRKF FDP D+VVD
Sbjct 240 VLNEESEGGARRRSWRKWLFPPSPSRCPAPPGPSESIVWEGFGPDSNRKFCFDPFDVVVD 299
Query 268 ESESGSLEVLYLCKVARFEDPRTGDASEHGHTTFFYNTVKAEGRMHYSRILPNLYVGSCP 327
+E G LE LY+C++A F DPR G E+ TT FYN+VK+E RMHYS I P +VGSCP
Sbjct 300 VNELGDLECLYICRIAHFRDPRAGGVGEYDLTTRFYNSVKSECRMHYSTIFPRFFVGSCP 359
Query 328 RQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLYESAGLRYVWMPT 387
RQLKHIR LKEELKVT V N+QT D+ N+PDP+AS R+ EA QLY+ +GLRYVW+PT
Sbjct 360 RQLKHIRHLKEELKVTCVVNLQTEQDLCNNYPDPIASSRSAEAVSQLYDGSGLRYVWLPT 419
Query 388 EDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAYLCFCVGLDVRKVN 447
DM D+ RK+AVA A +L L Q+GHSVY+HCNAGVGRSVAA A+LCF VGLD+RKVN
Sbjct 420 ADMCDSARKIAVANAAFLLLGLFQSGHSVYMHCNAGVGRSVAAACAFLCFAVGLDLRKVN 479
Query 448 FSVSTKRPV 456
F + +RPV
Sbjct 480 FLICARRPV 488
> mmu:13853 Epm2a; epilepsy, progressive myoclonic epilepsy, type
2 gene alpha (EC:3.1.3.16 3.1.3.48); K14165 dual specificity
phosphatase [EC:3.1.3.16 3.1.3.48]
Length=330
Score = 175 bits (444), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 96/225 (42%), Positives = 134/225 (59%), Gaps = 23/225 (10%)
Query 238 PSNSLHVIWEGSGPGTNRKFFFDPMDIVVDESESGSLEVLYLCKVARFEDPRTGDASEHG 297
P LH WEG+GP +R ++ ++V + +Y V + + TG +E
Sbjct 92 PGGELH--WEGNGPHHDRCCTYNEDNLV---------DGVYCLPVGHWIEA-TGHTNEMK 139
Query 298 HTTFFYNTVKAEGRMHYSRILPNLYVGSCPRQLKHIR-QLKEELKVTVVFNMQTADDMKT 356
HTT FY + MHYSRILPN+++GSCPRQL+H+ +LK EL VT V N QT D+
Sbjct 140 HTTDFYFNIAGHQAMHYSRILPNIWLGSCPRQLEHVTIKLKHELGVTAVMNFQTEWDIIQ 199
Query 357 N------FPDPLASKRTPEAAGQLYESAGLRYVWMPTEDMSDACRKLAVAQCALILDALI 410
N +P+P+ TP+ +LY+ GL Y+WMPT DMS R + Q +L AL+
Sbjct 200 NSSGCNRYPEPM----TPDTMMKLYKEEGLSYIWMPTPDMSTEGRVQMLPQAVCLLHALL 255
Query 411 QNGHSVYVHCNAGVGRSVAAVGAYLCFCVGLDVRKVNFSVSTKRP 455
+NGH+VYVHCNAGVGRS AAV +L + +G ++RKV + + KRP
Sbjct 256 ENGHTVYVHCNAGVGRSTAAVCGWLHYVIGWNLRKVQYFIMAKRP 300
Score = 31.6 bits (70), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 38/82 (46%), Gaps = 13/82 (15%)
Query 27 MRLRFSCVAFVPPG-----SQLGVSGGPDFLGRWDPCKCLPLLPHSSPSEGGI----EPS 77
M RF V VPP +L ++G LGRW+P + L P + + EP
Sbjct 1 MLFRFGVV--VPPAVAGARQELLLAGSRPELGRWEPHGAVRLRPAGTAAGAAALALQEPG 58
Query 78 LWFADVEVNEKSLERCGKSAAG 99
LW A+VE+ ++ E G + G
Sbjct 59 LWLAEVEL--EAYEEAGGAEPG 78
> xla:444115 epm2a, MGC80469; epilepsy, progressive myoclonus
type 2A, Lafora disease (laforin); K14165 dual specificity phosphatase
[EC:3.1.3.16 3.1.3.48]
Length=313
Score = 168 bits (425), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 90/219 (41%), Positives = 132/219 (60%), Gaps = 21/219 (9%)
Query 244 VIWEGSGPGTNRKFFFDPMDIVVDESESGSLEVLYLCKVARFEDPRTGDASEHGHTTFFY 303
+IWEG+GP +R +D +IV + +Y V + + TG +E HTT FY
Sbjct 79 LIWEGNGPHHDRCCVYDDSNIV---------DGVYCHPVGHWIET-TGHTNEMKHTTDFY 128
Query 304 NTVKAEGRMHYSRILPNLYVGSCPRQLKHIR-QLKEELKVTVVFNMQTADDMKTN----- 357
MH+SRILPN+++GSCPRQL+H+ ++K EL VT V N QT D+ N
Sbjct 129 FHFAGNQAMHFSRILPNIWLGSCPRQLEHVTVKMKHELGVTAVLNFQTEWDVIQNSSGCN 188
Query 358 -FPDPLASKRTPEAAGQLYESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSV 416
+P+P++ PE +LY+ G+ Y+W+PT DMS R + Q +L L++NGH+V
Sbjct 189 RYPEPMS----PETLFRLYKEVGITYIWIPTPDMSTEGRIRMLPQAVYLLFGLLENGHTV 244
Query 417 YVHCNAGVGRSVAAVGAYLCFCVGLDVRKVNFSVSTKRP 455
YVHCNAGVGRS AAV +L + +G +RKV + ++++RP
Sbjct 245 YVHCNAGVGRSTAAVCGFLMYVIGWSLRKVQYFLASRRP 283
> hsa:7957 EPM2A, EPM2, MELF; epilepsy, progressive myoclonus
type 2A, Lafora disease (laforin) (EC:3.1.3.16 3.1.3.48); K14165
dual specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=317
Score = 166 bits (419), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 89/217 (41%), Positives = 128/217 (58%), Gaps = 21/217 (9%)
Query 246 WEGSGPGTNRKFFFDPMDIVVDESESGSLEVLYLCKVARFEDPRTGDASEHGHTTFFYNT 305
WEG+GP +R ++ E+ ++ +Y + + + TG +E HTT FY
Sbjct 99 WEGNGPHHDRCCTYN---------ENNLVDGVYCLPIGHWIEA-TGHTNEMKHTTDFYFN 148
Query 306 VKAEGRMHYSRILPNLYVGSCPRQLKHIR-QLKEELKVTVVFNMQTADDMKTN------F 358
+ MHYSRILPN+++GSCPRQ++H+ +LK EL +T V N QT D+ N +
Sbjct 149 IAGHQAMHYSRILPNIWLGSCPRQVEHVTIKLKHELGITAVMNFQTEWDIVQNSSGCNRY 208
Query 359 PDPLASKRTPEAAGQLYESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYV 418
P+P+ TP+ +LY GL Y+WMPT DMS R + Q +L AL++ GH VYV
Sbjct 209 PEPM----TPDTMIKLYREEGLAYIWMPTPDMSTEGRVQMLPQAVCLLHALLEKGHIVYV 264
Query 419 HCNAGVGRSVAAVGAYLCFCVGLDVRKVNFSVSTKRP 455
HCNAGVGRS AAV +L + +G ++RKV + + KRP
Sbjct 265 HCNAGVGRSTAAVCGWLQYVMGWNLRKVQYFLMAKRP 301
> ath:AT3G10940 protein phosphatase-related
Length=282
Score = 55.5 bits (132), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 37/145 (25%), Positives = 71/145 (48%), Gaps = 8/145 (5%)
Query 312 MHYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAA 371
M+Y+ I L VGS P++ + I LK+E V + N+Q D++ D ++
Sbjct 91 MNYTLIRDELIVGSQPQKPEDIDHLKQEQNVAYILNLQQDKDIEYWGID-------LDSI 143
Query 372 GQLYESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHS-VYVHCNAGVGRSVAA 430
+ + G+R++ P +D + + + L+ + G VYVHC+AG+GR+
Sbjct 144 VRRCKELGIRHMRRPAKDFDPLSLRSQLPKAVSSLEWAVSEGKGRVYVHCSAGLGRAPGV 203
Query 431 VGAYLCFCVGLDVRKVNFSVSTKRP 455
AY+ + +++ ++ +KRP
Sbjct 204 SIAYMYWFCDMNLNTAYDTLVSKRP 228
> tgo:TGME49_108830 dual specificity phosphatase, catalytic domain
containing protein (EC:3.1.3.16); K14819 dual specificity
phosphatase 12 [EC:3.1.3.16 3.1.3.48]
Length=172
Score = 47.0 bits (110), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 37/79 (46%), Gaps = 0/79 (0%)
Query 376 ESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAYL 435
E GL Y + ED S L + +D + +V VHC AGV RS + V +YL
Sbjct 44 EREGLDYFRVDVEDTSREPLHLYFEEAGQFIDRYVSRQQTVLVHCKAGVSRSASVVLSYL 103
Query 436 CFCVGLDVRKVNFSVSTKR 454
C +++ F V TKR
Sbjct 104 IGCKKFALQEAFFHVLTKR 122
> mmu:240672 Dusp5, Gm337; dual specificity phosphatase 5; K04459
dual specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=384
Score = 45.1 bits (105), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 40/140 (28%), Positives = 56/140 (40%), Gaps = 22/140 (15%)
Query 317 ILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLYE 376
ILP LY+GS K + L +T + N+ S+RT EA
Sbjct 182 ILPFLYLGSAYHASKC--EFLANLHITALLNV---------------SRRTSEAC----- 219
Query 377 SAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAYLC 436
+ L Y W+P ED A + +D + + G V VHC AGV RS AYL
Sbjct 220 TTHLHYKWIPVEDSHTADISSHFQEAIDFIDCVREEGGKVLVHCEAGVSRSPTICMAYLM 279
Query 437 FCVGLDVRKVNFSVSTKRPV 456
+++ V +R V
Sbjct 280 KTKQFRLKEAFDYVKQRRSV 299
> ath:AT3G52180 SEX4; SEX4 (STARCH-EXCESS 4); polysaccharide binding
/ protein tyrosine/serine/threonine phosphatase
Length=292
Score = 44.7 bits (104), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 37/151 (24%), Positives = 75/151 (49%), Gaps = 8/151 (5%)
Query 305 TVKAEGRMHYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLAS 364
T + E M+Y+ I P+L VGSC + + + +L++ + V +F +Q D++ D
Sbjct 89 TYRHELGMNYNFIRPDLIVGSCLQTPEDVDKLRK-IGVKTIFCLQQDPDLEYFGVDI--- 144
Query 365 KRTPEAAGQLYESAGLRYVWMPTEDMSDACRKLAV-AQCALILDALIQNGHSVYVHCNAG 423
+ +A + Y + ++++ D ++ + A + A+ +NG YVHC AG
Sbjct 145 -SSIQAYAKKY--SDIQHIRCEIRDFDAFDLRMRLPAVVGTLYKAVKRNGGVTYVHCTAG 201
Query 424 VGRSVAAVGAYLCFCVGLDVRKVNFSVSTKR 454
+GR+ A Y+ + G + + + + +KR
Sbjct 202 MGRAPAVALTYMFWVQGYKLMEAHKLLMSKR 232
> ath:AT3G01510 5'-AMP-activated protein kinase beta-1 subunit-related
Length=591
Score = 44.3 bits (103), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 55/124 (44%), Gaps = 8/124 (6%)
Query 312 MHYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAA 371
M YS+I +YVGSC + + + L E +T + N Q + + D ++ A
Sbjct 290 MRYSKITEQIYVGSCIQTEEDVENLSEA-GITAILNFQGGTEAQNWGID----SQSINDA 344
Query 372 GQLYESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAV 431
Q E + Y P +D + + C +L L++ H V+V C G RS A V
Sbjct 345 CQKSEVLMINY---PIKDADSFDLRKKLPLCVGLLLRLLKKNHRVFVTCTTGFDRSSACV 401
Query 432 GAYL 435
AYL
Sbjct 402 IAYL 405
> dre:100000665 dual specificity phosphatase 3-like; K04459 dual
specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=200
Score = 44.3 bits (103), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 36/146 (24%), Positives = 57/146 (39%), Gaps = 16/146 (10%)
Query 313 HYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADD-MKTNFPDPLASKRTPEAA 371
H++ + P +Y+G+ +R + L VT + N+ + M N
Sbjct 43 HFNEVFPRIYIGNAFVAQNVMRL--QRLGVTHILNVAEGNSFMHVN------------TN 88
Query 372 GQLYESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGH-SVYVHCNAGVGRSVAA 430
+ Y G+ Y + D + A +D + +G VYVHC G RS
Sbjct 89 AEFYAGTGITYHGIQANDTEQFNISAFFEEAADFIDKALAHGKGKVYVHCREGYSRSPTI 148
Query 431 VGAYLCFCVGLDVRKVNFSVSTKRPV 456
V AYL +DVR +V KR +
Sbjct 149 VIAYLMLRHKMDVRVATATVRHKREI 174
> hsa:1847 DUSP5, DUSP, HVH3; dual specificity phosphatase 5 (EC:3.1.3.16
3.1.3.48); K04459 dual specificity phosphatase [EC:3.1.3.16
3.1.3.48]
Length=384
Score = 43.9 bits (102), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 49/119 (41%), Gaps = 22/119 (18%)
Query 317 ILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLYE 376
ILP LY+GS K + L +T + N+ S+RT EA
Sbjct 182 ILPFLYLGSAYHASKC--EFLANLHITALLNV---------------SRRTSEAC----- 219
Query 377 SAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAYL 435
+ L Y W+P ED A + +D + + G V VHC AG+ RS AYL
Sbjct 220 ATHLHYKWIPVEDSHTADISSHFQEAIDFIDCVREKGGKVLVHCEAGISRSPTICMAYL 278
> mmu:80915 Dusp12, 1190004O14Rik, AA027408, AW049275, ESTM36,
LMW-DSP4, MGC117646, T-DSP4, VH1, mVH1; dual specificity phosphatase
12 (EC:3.1.3.16 3.1.3.48); K14819 dual specificity
phosphatase 12 [EC:3.1.3.16 3.1.3.48]
Length=339
Score = 42.7 bits (99), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 40/145 (27%), Positives = 61/145 (42%), Gaps = 23/145 (15%)
Query 313 HYSRILPNLYVGSCPRQLK--HIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEA 370
H + P LY+G + H+R E +T V + D + FP
Sbjct 26 HAVEVRPGLYLGGAAAVAEPGHLR----EAGITAVLTV----DSEPAFP----------- 66
Query 371 AGQLYESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAA 430
AG +E GLR +++P D + + +C + G +V VHC+AGV RSVA
Sbjct 67 AGAGFE--GLRSLFVPALDKPETDLLSHLDRCVAFIGQARSEGRAVLVHCHAGVSRSVAV 124
Query 431 VGAYLCFCVGLDVRKVNFSVSTKRP 455
V A++ L K + T +P
Sbjct 125 VMAFIMKTDQLTFEKAYDILRTVKP 149
> mmu:245857 Ssh3, BC028922, SSH-3, mSSH-3L; slingshot homolog
3 (Drosophila) (EC:3.1.3.16 3.1.3.48); K05766 slingshot
Length=649
Score = 41.6 bits (96), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 44/144 (30%), Positives = 62/144 (43%), Gaps = 26/144 (18%)
Query 315 SRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLA--SKRTPEAAG 372
SRI P+LY+GS ++ +L++ +V+ + NM A ++ FP+ + R +
Sbjct 327 SRIFPHLYLGS-EWNAANLEELQKN-RVSHILNM--AREIDNFFPERFTYYNVRVWDE-- 380
Query 373 QLYESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVG 432
ESA L W T I DA Q G V VHC GV RS A V
Sbjct 381 ---ESAQLLPHWKETH--------------RFIEDARAQ-GTRVLVHCKMGVSRSAATVL 422
Query 433 AYLCFCVGLDVRKVNFSVSTKRPV 456
AY G D+ + V RP+
Sbjct 423 AYAMKQYGWDLEQALIHVQELRPI 446
> xla:444752 dusp5, MGC84792; dual specificity phosphatase 5;
K04459 dual specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=373
Score = 41.2 bits (95), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 33/119 (27%), Positives = 49/119 (41%), Gaps = 21/119 (17%)
Query 317 ILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLYE 376
ILP LY+GS K + L +T + N+ + K +P+ + Y
Sbjct 173 ILPFLYLGSAYHASKC--EFLANLHITALLNV--------------SRKSSPDFCKEQYS 216
Query 377 SAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAYL 435
Y W+P ED A + +D++ + G V VHC AG+ RS AYL
Sbjct 217 -----YKWIPVEDNHTADISSHFQEAIDFIDSVKRAGGRVLVHCEAGISRSPTICMAYL 270
> dre:554088 styx, im:6908851, si:dkey-119m7.5, zgc:110621; serine/threonine/tyrosine
interacting protein
Length=224
Score = 40.8 bits (94), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 36/146 (24%), Positives = 59/146 (40%), Gaps = 19/146 (13%)
Query 311 RMHYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEA 370
R ILP L++G +K + E+ +T + ++ D++ NF P +
Sbjct 27 RREMQEILPGLFLGPYSAAMKSKLSMLEKQGITHIVCVR--QDIEANFIKPNFPHK---- 80
Query 371 AGQLYESAGLRYVWMPTED--MSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSV 428
RY+ + D + + R + +D ++ G V VH NAG+ RS
Sbjct 81 ---------FRYLVLDIADNPVENIIRYFPTTKE--FIDGCLETGGKVLVHGNAGISRSA 129
Query 429 AAVGAYLCFCVGLDVRKVNFSVSTKR 454
A V AYL G+ R V +R
Sbjct 130 ALVIAYLMETFGVKYRDAFSHVQERR 155
> dre:568344 dusp3, si:bz18k17.3, si:rp71-18k17.3; dual specificity
phosphatase 3 (vaccinia virus phosphatase VH1-related);
K14165 dual specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=177
Score = 40.4 bits (93), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 35/144 (24%), Positives = 55/144 (38%), Gaps = 15/144 (10%)
Query 314 YSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNM-QTADDMKTNFPDPLASKRTPEAAG 372
+ + P + +G+ R L EL VT + N + DM N
Sbjct 29 FHEVYPGILLGNESAATNVTRLL--ELGVTHILNAAEGQSDMHVN------------TDA 74
Query 373 QLYESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVG 432
+ Y G+ Y +P D + + + + ++ VYVHC G RS A V
Sbjct 75 EYYADTGIIYHGIPAFDTDHFDLSIYFEEASDFIQRALEMKGKVYVHCQKGYSRSAALVI 134
Query 433 AYLCFCVGLDVRKVNFSVSTKRPV 456
A+L +DVR +V KR +
Sbjct 135 AHLMLQHNMDVRAAVATVREKREI 158
> hsa:78986 DUSP26, DUSP24, LDP-4, MGC1136, MGC2627, MKP8, NATA1,
SKRP3; dual specificity phosphatase 26 (putative) (EC:3.1.3.16
3.1.3.48); K14165 dual specificity phosphatase [EC:3.1.3.16
3.1.3.48]
Length=211
Score = 40.0 bits (92), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 43/149 (28%), Positives = 60/149 (40%), Gaps = 26/149 (17%)
Query 313 HYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAG 372
H + P LY+G + + + R+ L +T V N + + TPEA
Sbjct 61 HADEVWPGLYLGD--QDMANNRRELRRLGITHVLNASHSR-----------WRGTPEA-- 105
Query 373 QLYESAGLRYVWM-----PTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRS 427
YE G+RY+ + P DMS + A I AL Q G + VHC GV RS
Sbjct 106 --YEGLGIRYLGVEAHDSPAFDMSIHFQTAA----DFIHRALSQPGGKILVHCAVGVSRS 159
Query 428 VAAVGAYLCFCVGLDVRKVNFSVSTKRPV 456
V AYL L + + V R +
Sbjct 160 ATLVLAYLMLYHHLTLVEAIKKVKDHRGI 188
> mmu:75590 Dusp9, Dusp4, Mpk4, Pyst3; dual specificity phosphatase
9; K04459 dual specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=452
Score = 40.0 bits (92), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 37/121 (30%), Positives = 50/121 (41%), Gaps = 22/121 (18%)
Query 316 RILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLY 375
+ILPNLY+GS R ++ L + L + + N+ TP
Sbjct 274 QILPNLYLGSA-RDSANLESLAK-LGIRYILNV------------------TPNLPNLFE 313
Query 376 ESAGLRYVWMPTEDM-SDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAY 434
++ Y +P D S + A I +AL QN V VHC AGV RSV AY
Sbjct 314 KNGDFHYKQIPISDHWSQNLSQFFPEAIAFIDEALSQNC-GVLVHCLAGVSRSVTVTVAY 372
Query 435 L 435
L
Sbjct 373 L 373
> mmu:56291 Styx, 0610039A20Rik, MGC106795, hStyxb; serine/threonine/tyrosine
interaction protein
Length=223
Score = 40.0 bits (92), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 36/144 (25%), Positives = 61/144 (42%), Gaps = 15/144 (10%)
Query 311 RMHYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEA 370
R +LP L++G +K + ++ +T + ++ +++ NF P
Sbjct 26 RREMQEVLPGLFLGPYSSAMKSKLPILQKHGITHIICIR--QNIEANFIKP--------N 75
Query 371 AGQLYESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAA 430
QL+ L P E++ R + + +D +QNG V VH NAG+ RS A
Sbjct 76 FQQLFRYLVLDIADNPVENI---IRFFPMTKE--FIDGSLQNGGKVLVHGNAGISRSAAF 130
Query 431 VGAYLCFCVGLDVRKVNFSVSTKR 454
V AY+ G+ R V +R
Sbjct 131 VIAYIMETFGMKYRDAFAYVQERR 154
> mmu:100040244 Gm14698, OTTMUSG00000017743; predicted gene 14698
Length=223
Score = 40.0 bits (92), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 36/144 (25%), Positives = 61/144 (42%), Gaps = 15/144 (10%)
Query 311 RMHYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEA 370
R +LP L++G +K + ++ +T + ++ +++ NF P
Sbjct 26 RREMQEVLPGLFLGPYSSAMKSKLPILQKHGITHIICIR--QNIEANFIKP--------N 75
Query 371 AGQLYESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAA 430
QL+ L P E++ R + + +D +QNG V VH NAG+ RS A
Sbjct 76 FQQLFRYLVLDIADNPVENI---IRFFPMTKE--FIDGSLQNGGKVLVHGNAGISRSAAF 130
Query 431 VGAYLCFCVGLDVRKVNFSVSTKR 454
V AY+ G+ R V +R
Sbjct 131 VIAYIMETFGMKYRDAFAYVQERR 154
> ath:AT3G23610 dual specificity protein phosphatase (DsPTP1);
K05766 slingshot
Length=201
Score = 39.7 bits (91), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 35/75 (46%), Gaps = 0/75 (0%)
Query 382 YVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAYLCFCVGL 441
Y + D D ++ +C +D + G SV VHC G RSV V AYL G+
Sbjct 97 YKVVRVVDKEDTNLEMYFDECVDFIDEAKRQGGSVLVHCFVGKSRSVTIVVAYLMKKHGM 156
Query 442 DVRKVNFSVSTKRPV 456
+ + V +KRPV
Sbjct 157 TLAQALQHVKSKRPV 171
> mmu:66959 Dusp26, 2310043K02Rik, Skrp3; dual specificity phosphatase
26 (putative) (EC:3.1.3.16 3.1.3.48); K14165 dual specificity
phosphatase [EC:3.1.3.16 3.1.3.48]
Length=211
Score = 39.7 bits (91), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 50/126 (39%), Gaps = 18/126 (14%)
Query 313 HYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAG 372
H + P LY+G + + + R+ L +T V N + TPEA
Sbjct 61 HADEVWPGLYLGD--QDMANNRRELRRLGITHVLNASHNR-----------WRGTPEA-- 105
Query 373 QLYESAGLRYVWMPTEDMSDACRKLAVAQCA-LILDALIQNGHSVYVHCNAGVGRSVAAV 431
YE G+RY+ + D + A I AL Q G + VHC GV RS V
Sbjct 106 --YEGLGIRYLGVEAHDSPAFDMSIHFQTAADFIHRALSQPGGKILVHCAVGVSRSATLV 163
Query 432 GAYLCF 437
AYL
Sbjct 164 LAYLML 169
> mmu:252864 Dusp15, AI851682, LMW-DSP10, T-DSP10; dual specificity
phosphatase-like 15 (EC:3.1.3.16 3.1.3.48); K14165 dual
specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=235
Score = 39.7 bits (91), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 59/142 (41%), Gaps = 23/142 (16%)
Query 315 SRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQL 374
+++LP LY+G+ K QL K+T + ++ + P PL T
Sbjct 6 TKVLPGLYLGNFI-DAKDPDQLGRN-KITHIISIHES-------PQPLLQDIT------- 49
Query 375 YESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAY 434
Y+ + D + K +C + + NG + VHC AG+ RS V AY
Sbjct 50 -------YLRISVSDTPEVPIKKHFKECVHFIHSCRLNGGNCLVHCFAGISRSTTIVIAY 102
Query 435 LCFCVGLDVRKVNFSVSTKRPV 456
+ GL ++V ++ RP+
Sbjct 103 VMTVTGLGWQEVLEAIKASRPI 124
> sce:YFR028C CDC14, OAF3; Cdc14p (EC:3.1.3.48); K06639 cell division
cycle 14 [EC:3.1.3.48]
Length=551
Score = 39.3 bits (90), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 39/68 (57%), Gaps = 3/68 (4%)
Query 375 YESAGLRYVWMPTEDMSDACRKLAVAQCAL-ILDALIQNGHSVYVHCNAGVGRSVAAVGA 433
+E G++++ + ED + C L++ + + + +I+ G + VHC AG+GR+ +GA
Sbjct 239 FEDIGIQHLDLIFEDGT--CPDLSIVKNFVGAAETIIKRGGKIAVHCKAGLGRTGCLIGA 296
Query 434 YLCFCVGL 441
+L + G
Sbjct 297 HLIYTYGF 304
> tgo:TGME49_021500 dual specificity protein phosphatase, catalytic
domain-containing protein (EC:3.1.3.16)
Length=982
Score = 39.3 bits (90), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 26/53 (49%), Gaps = 0/53 (0%)
Query 404 LILDALIQNGHSVYVHCNAGVGRSVAAVGAYLCFCVGLDVRKVNFSVSTKRPV 456
L+ I+ +V VHCN GV RSVA V +YL + V + RPV
Sbjct 121 LMERVRIRESGNVLVHCNQGVSRSVAMVASYLIRFFKMSVEDAVALIQVNRPV 173
> xla:495329 styx-b; serine/threonine/tyrosine interacting protein
Length=223
Score = 39.3 bits (90), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 36/146 (24%), Positives = 62/146 (42%), Gaps = 19/146 (13%)
Query 311 RMHYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEA 370
R ILP L++G +K + ++ +T V ++ +++ NF P
Sbjct 26 RREMQEILPGLFLGPYSAAMKSKLSVLQKCGITHVICIR--QNIEANFIKP--------N 75
Query 371 AGQLYESAGLRYVWMPTED--MSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSV 428
QL+ RY+ + D + + R ++ +D +Q G V +H NAG+ RS
Sbjct 76 FQQLF-----RYLVLDIADNPVENIIRFFLTSK--EFIDGCLQTGGKVLIHGNAGISRSA 128
Query 429 AAVGAYLCFCVGLDVRKVNFSVSTKR 454
A V AY+ G+ R V +R
Sbjct 129 ALVIAYIMETFGIKYRDAFTYVQERR 154
> xla:734588 styx-a, MGC115529, styx; serine/threonine/tyrosine
interacting protein
Length=223
Score = 38.9 bits (89), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 36/144 (25%), Positives = 61/144 (42%), Gaps = 15/144 (10%)
Query 311 RMHYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEA 370
R ILP L++G +K + ++ +T V ++ +++ NF P
Sbjct 26 RREMQEILPGLFLGPYSAAMKSKLSVFQKCGITHVICIR--QNIEANFIKP--------N 75
Query 371 AGQLYESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAA 430
QL+ L P E++ R +++ +D +Q G V +H NAG+ RS A
Sbjct 76 FQQLFRYLVLDIADNPVENI---IRFFPMSK--EFIDGCLQTGGKVLIHGNAGISRSAAL 130
Query 431 VGAYLCFCVGLDVRKVNFSVSTKR 454
V AY+ G+ R V +R
Sbjct 131 VIAYIMETFGIKYRDAFTYVQERR 154
> dre:558079 dual specificity phosphatase 8-like
Length=462
Score = 38.9 bits (89), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 29/58 (50%), Gaps = 0/58 (0%)
Query 399 VAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAYLCFCVGLDVRKVNFSVSTKRPV 456
+ Q +D + G SV VHC AG+ RS A AY+ + + +D+ V +RP
Sbjct 77 IPQALHFIDGAMSAGCSVLVHCAAGISRSPALAVAYVMYSLKMDLDHAYRFVKERRPT 134
> dre:100004731 novel protein similar to vertebrate dual specificity
phosphatase and pro isomerase domain containing 1 (DUPD1);
K14165 dual specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=186
Score = 38.5 bits (88), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 38/142 (26%), Positives = 52/142 (36%), Gaps = 15/142 (10%)
Query 313 HYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAG 372
H + PNL++G + H R L VT V N + K + G
Sbjct 34 HVDEVWPNLFLGD--MYMSHDRYGLWSLGVTHVLNAAHG---------KMCCKGNDDYYG 82
Query 373 QLYESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVG 432
+ G+ +PT D+S I DAL G V+VHC G+ RS A V
Sbjct 83 TTVKYYGVPANDLPTFDIS----PFFYPSAQYIHDALSTTGAKVFVHCAVGMSRSAALVL 138
Query 433 AYLCFCVGLDVRKVNFSVSTKR 454
AYL + V +R
Sbjct 139 AYLMIYCNFSLVDAILKVKERR 160
> cel:F28C6.8 hypothetical protein
Length=150
Score = 38.5 bits (88), Expect = 0.056, Method: Composition-based stats.
Identities = 15/61 (24%), Positives = 31/61 (50%), Gaps = 0/61 (0%)
Query 375 YESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAY 434
+++ G+ + +P +D + + + + ++++ G +VYVHC AG RS Y
Sbjct 40 WKNEGVEFFAVPMKDFTGTAPRAEINEAVEFIESVASKGKTVYVHCKAGRTRSATVATCY 99
Query 435 L 435
L
Sbjct 100 L 100
> xla:398829 dusp6, MGC68682, mkp3, pyst1; dual specificity phosphatase
6; K04459 dual specificity phosphatase [EC:3.1.3.16
3.1.3.48]
Length=378
Score = 37.4 bits (85), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 57/137 (41%), Gaps = 29/137 (21%)
Query 316 RILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLY 375
ILP LY+G C + ++ L+E F ++ ++ N P+ L+
Sbjct 206 EILPYLYLG-CAKDSTNLDVLEE-------FGIKYILNVTPNLPN-------------LF 244
Query 376 ESAG-LRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAY 434
E+AG RY +P D + +D V VHC AG+ RSV AY
Sbjct 245 ENAGEFRYKQIPISDHWSQNLSQFFPEAISFIDEARGKSCGVLVHCLAGISRSVTVTVAY 304
Query 435 LCFCVGLDVRKVNFSVS 451
L ++K+N S++
Sbjct 305 L-------MQKLNLSMN 314
> hsa:1852 DUSP9, MKP-4, MKP4; dual specificity phosphatase 9
(EC:3.1.3.16 3.1.3.48); K04459 dual specificity phosphatase
[EC:3.1.3.16 3.1.3.48]
Length=384
Score = 37.4 bits (85), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 36/121 (29%), Positives = 49/121 (40%), Gaps = 22/121 (18%)
Query 316 RILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLY 375
+ILPNLY+GS R ++ L + L + + N+ TP
Sbjct 206 QILPNLYLGSA-RDSANLESLAK-LGIRYILNV------------------TPNLPNFFE 245
Query 376 ESAGLRYVWMPTEDM-SDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAY 434
++ Y +P D S + I +AL QN V VHC AGV RSV AY
Sbjct 246 KNGDFHYKQIPISDHWSQNLSRFFPEAIEFIDEALSQNC-GVLVHCLAGVSRSVTVTVAY 304
Query 435 L 435
L
Sbjct 305 L 305
> xla:734809 dusp18, MGC131167; dual specificity phosphatase 18;
K14165 dual specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=191
Score = 37.4 bits (85), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 36/81 (44%), Gaps = 0/81 (0%)
Query 376 ESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAYL 435
E+ L YV +P D D C A + + G + +HC AG+ RS A+L
Sbjct 63 EAPELEYVNIPVSDTPDTCLLQYFEDIADKIHTVKAGGGNTLLHCVAGISRSPTLCLAFL 122
Query 436 CFCVGLDVRKVNFSVSTKRPV 456
GL ++ + V T RP+
Sbjct 123 MKYHGLSLQAAHDWVKTCRPI 143
> hsa:6815 STYX, FLJ42934; serine/threonine/tyrosine interacting
protein
Length=223
Score = 37.4 bits (85), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 36/146 (24%), Positives = 62/146 (42%), Gaps = 19/146 (13%)
Query 311 RMHYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEA 370
R ILP L++G +K + ++ +T + ++ +++ NF P
Sbjct 26 RREMQEILPGLFLGPYSSAMKSKLPVLQKHGITHIICIR--QNIEANFIKP--------N 75
Query 371 AGQLYESAGLRYVWMPTED--MSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSV 428
QL+ RY+ + D + + R + + +D +Q G V VH NAG+ RS
Sbjct 76 FQQLF-----RYLVLDIADNPVENIIRFFPMTKE--FIDGSLQMGGKVLVHGNAGISRSA 128
Query 429 AAVGAYLCFCVGLDVRKVNFSVSTKR 454
A V AY+ G+ R V +R
Sbjct 129 AFVIAYIMETFGMKYRDAFAYVQERR 154
> hsa:100290651 serine/threonine/tyrosine-interacting protein-like
Length=223
Score = 37.4 bits (85), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 36/146 (24%), Positives = 62/146 (42%), Gaps = 19/146 (13%)
Query 311 RMHYSRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEA 370
R ILP L++G +K + ++ +T + ++ +++ NF P
Sbjct 26 RREMQEILPGLFLGPYSSAMKSKLPVLQKHGITHIICIR--QNIEANFIKP--------N 75
Query 371 AGQLYESAGLRYVWMPTED--MSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSV 428
QL+ RY+ + D + + R + + +D +Q G V VH NAG+ RS
Sbjct 76 FQQLF-----RYLVLDIADNPVENIIRFFPMTKE--FIDGSLQMGGKVLVHGNAGISRSA 128
Query 429 AAVGAYLCFCVGLDVRKVNFSVSTKR 454
A V AY+ G+ R V +R
Sbjct 129 AFVIAYIMETFGMKYRDAFAYVQERR 154
> dre:445057 dusp2, wu:fj40g04, zgc:91929; dual specificity phosphatase
2; K04459 dual specificity phosphatase [EC:3.1.3.16
3.1.3.48]
Length=333
Score = 37.4 bits (85), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 50/119 (42%), Gaps = 22/119 (18%)
Query 317 ILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLYE 376
ILP L++GS + R+ E +T V N+ ++ L+E
Sbjct 182 ILPFLFLGSAHHSSR--RETLERNGITAVLNVSSS-------------------CPNLFE 220
Query 377 SAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAYL 435
L+Y + ED A ++ + +D++ + G V VHC AG+ RS AYL
Sbjct 221 EE-LQYKTLKVEDSLAADIRVLFPEAIHFIDSIKEGGGRVLVHCQAGISRSATICLAYL 278
> dre:114436 dusp5, wu:fc03b11, wu:fc86e02; dual specificity phosphatase
5; K04459 dual specificity phosphatase [EC:3.1.3.16
3.1.3.48]
Length=368
Score = 37.4 bits (85), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 46/119 (38%), Gaps = 22/119 (18%)
Query 317 ILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLYE 376
ILP LY+GS + + +L +T + N+ D + A GQ
Sbjct 175 ILPFLYLGSAYHACR--QDYLSDLHITALLNVSRRD--------------SRPARGQY-- 216
Query 377 SAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAYL 435
Y W+P ED A + ++ + G V VHC AG+ RS AY+
Sbjct 217 ----NYKWIPVEDSHTADISSHFQEAIDFIERVKAEGGKVLVHCEAGISRSPTICMAYI 271
> xla:495060 hypothetical LOC495060; K04459 dual specificity phosphatase
[EC:3.1.3.16 3.1.3.48]
Length=379
Score = 37.4 bits (85), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 36/136 (26%), Positives = 57/136 (41%), Gaps = 29/136 (21%)
Query 317 ILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLYE 376
ILP LY+G C + ++ L+E F ++ ++ N P+ L+E
Sbjct 208 ILPYLYLG-CAKDSTNLDVLEE-------FGIKYILNVTPNLPN-------------LFE 246
Query 377 SAG-LRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAYL 435
+AG RY +P D + +D V VHC AG+ RSV AYL
Sbjct 247 NAGEFRYKQIPISDHWSQNLSQFFPEAISFIDEARGKSCGVLVHCLAGISRSVTVTVAYL 306
Query 436 CFCVGLDVRKVNFSVS 451
++K+N S++
Sbjct 307 -------MQKLNLSMN 315
> sce:YIR026C YVH1; Protein phosphatase involved in vegetative
growth at low temperatures, sporulation, and glycogen accumulation;
mutants are defective in 60S ribosome assembly; member
of the dual-specificity family of protein phosphatases (EC:3.1.3.48);
K14819 dual specificity phosphatase 12 [EC:3.1.3.16
3.1.3.48]
Length=364
Score = 37.4 bits (85), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 415 SVYVHCNAGVGRSVAAVGAYLCFCVGLDVRKVNFSVSTKRP 455
+V+ HC AG+ RSV + AYL + GL + +V K+P
Sbjct 112 AVFAHCQAGLSRSVTFIVAYLMYRYGLSLSMAMHAVKRKKP 152
> mmu:72349 Dusp3, 2210015O03Rik, 5031436O03Rik, T-DSP11, VHR;
dual specificity phosphatase 3 (vaccinia virus phosphatase
VH1-related) (EC:3.1.3.16 3.1.3.48); K04459 dual specificity
phosphatase [EC:3.1.3.16 3.1.3.48]
Length=185
Score = 37.0 bits (84), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 36/144 (25%), Positives = 57/144 (39%), Gaps = 16/144 (11%)
Query 315 SRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADD-MKTNFPDPLASKRTPEAAGQ 373
+ ++P +YVG+ + I QL ++L +T V N M N +
Sbjct 31 NEVVPRVYVGNASVA-QDITQL-QKLGITHVLNAAEGRSFMHVN------------TSAS 76
Query 374 LYESAGLRYVWMPTEDMSDACRKLAVAQCALILD-ALIQNGHSVYVHCNAGVGRSVAAVG 432
YE +G+ Y+ + D + + +D AL V VHC G RS V
Sbjct 77 FYEDSGITYLGIKANDTQEFNLSAYFERATDFIDQALAHKNGRVLVHCREGYSRSPTLVI 136
Query 433 AYLCFCVGLDVRKVNFSVSTKRPV 456
AYL +DV+ +V R +
Sbjct 137 AYLMMRQKMDVKSALSTVRQNREI 160
> ath:AT3G06110 MKP2; MKP2 (MAPK PHOSPHATASE 2); MAP kinase phosphatase/
protein tyrosine/serine/threonine phosphatase
Length=157
Score = 37.0 bits (84), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 401 QCALILDALIQNGHSVYVHCNAGVGRSVAAVGAYLCFCVGLDVRKVNFSVSTKR 454
+C +D IQ+G V VHC G+ RSV V AYL G+ K V ++R
Sbjct 80 ECYSFIDQAIQSGGGVLVHCFMGMSRSVTIVVAYLMKKHGMGFSKAMELVRSRR 133
> cel:Y54F10BM.13 hypothetical protein; K14165 dual specificity
phosphatase [EC:3.1.3.16 3.1.3.48]
Length=227
Score = 37.0 bits (84), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 406 LDALIQNGHSVYVHCNAGVGRSVAAVGAYL 435
+D + QN V++HCNAG+ RS V AYL
Sbjct 153 IDKVRQNEGIVFIHCNAGISRSATFVVAYL 182
> hsa:138639 PTPDC1, FLJ42922, PTP9Q22; protein tyrosine phosphatase
domain containing 1 (EC:3.1.3.48); K01104 protein-tyrosine
phosphatase [EC:3.1.3.48]
Length=806
Score = 37.0 bits (84), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 31/67 (46%), Gaps = 7/67 (10%)
Query 395 RKLAVAQCALILDAL------IQNGHSVYVHCNAGVGRSVAAVGAYLCFCVGLDVRKVNF 448
+ VA ILD + +Q G V +HC+AG+GR+ + YL F + +
Sbjct 212 KDYGVASLTTILDMVKVMTFALQEG-KVAIHCHAGLGRTGVLIACYLVFATRMTADQAII 270
Query 449 SVSTKRP 455
V KRP
Sbjct 271 FVRAKRP 277
> mmu:218232 Ptpdc1, AI843923, AW456874, Naa-1; protein tyrosine
phosphatase domain containing 1 (EC:3.1.3.48); K01104 protein-tyrosine
phosphatase [EC:3.1.3.48]
Length=747
Score = 37.0 bits (84), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 31/67 (46%), Gaps = 7/67 (10%)
Query 395 RKLAVAQCALILDAL------IQNGHSVYVHCNAGVGRSVAAVGAYLCFCVGLDVRKVNF 448
+ VA ILD + +Q G V VHC+AG+GR+ + YL F + +
Sbjct 160 KDYGVASLTAILDMVKVMTFALQEG-KVAVHCHAGLGRTGVLIACYLVFATRMTADQAII 218
Query 449 SVSTKRP 455
V KRP
Sbjct 219 FVRAKRP 225
> hsa:1848 DUSP6, MKP3, PYST1; dual specificity phosphatase 6
(EC:3.1.3.16 3.1.3.48); K04459 dual specificity phosphatase
[EC:3.1.3.16 3.1.3.48]
Length=381
Score = 36.6 bits (83), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 35/137 (25%), Positives = 57/137 (41%), Gaps = 29/137 (21%)
Query 316 RILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLY 375
ILP LY+G C + ++ L+E F ++ ++ N P+ L+
Sbjct 209 EILPFLYLG-CAKDSTNLDVLEE-------FGIKYILNVTPNLPN-------------LF 247
Query 376 ESAG-LRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAY 434
E+AG +Y +P D + +D V VHC AG+ RSV AY
Sbjct 248 ENAGEFKYKQIPISDHWSQNLSQFFPEAISFIDEARGKNCGVLVHCLAGISRSVTVTVAY 307
Query 435 LCFCVGLDVRKVNFSVS 451
L ++K+N S++
Sbjct 308 L-------MQKLNLSMN 317
> dre:406340 dusp1, cb269, id:ibd2586, sb:cb269, wu:fd19g02, wu:fe50g09,
zgc:55920; dual specificity phosphatase 1; K04459
dual specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=360
Score = 36.6 bits (83), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 31/120 (25%), Positives = 47/120 (39%), Gaps = 22/120 (18%)
Query 316 RILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLY 375
ILP LY+GS + + + + L +T + N+ S P Y
Sbjct 178 EILPFLYLGSAYHASR--KDMLDMLGITALINV---------------SSNCPNHFEDHY 220
Query 376 ESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAYL 435
+ Y +P ED A + +D++ G V+VHC AG+ RS AYL
Sbjct 221 Q-----YKSIPVEDNHKANISSWFNEAIEFIDSVRNKGGRVFVHCQAGISRSATICLAYL 275
> xla:100505429 hypothetical protein LOC100505429; K04459 dual
specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=381
Score = 36.6 bits (83), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 35/137 (25%), Positives = 57/137 (41%), Gaps = 29/137 (21%)
Query 316 RILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLY 375
ILP LY+G C + ++ L+E F ++ ++ N P+ L+
Sbjct 209 EILPFLYLG-CAKDSTNLDVLEE-------FGIKYILNVTPNLPN-------------LF 247
Query 376 ESAG-LRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAY 434
E+AG +Y +P D + +D V VHC AG+ RSV AY
Sbjct 248 ENAGEFKYKQIPISDHWSQNLSQFFPEAISFIDEARGKNCGVLVHCLAGISRSVTVTVAY 307
Query 435 LCFCVGLDVRKVNFSVS 451
L ++K+N S++
Sbjct 308 L-------MQKLNLSMN 317
> mmu:67603 Dusp6, 1300019I03Rik, MGC98540, MKP-3, MKP3, PYST1;
dual specificity phosphatase 6 (EC:3.1.3.16 3.1.3.48); K04459
dual specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=381
Score = 36.6 bits (83), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 35/137 (25%), Positives = 57/137 (41%), Gaps = 29/137 (21%)
Query 316 RILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQLY 375
ILP LY+G C + ++ L+E F ++ ++ N P+ L+
Sbjct 209 EILPFLYLG-CAKDSTNLDVLEE-------FGIKYILNVTPNLPN-------------LF 247
Query 376 ESAG-LRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVGAY 434
E+AG +Y +P D + +D V VHC AG+ RSV AY
Sbjct 248 ENAGEFKYKQIPISDHWSQNLSQFFPEAISFIDEARGKNCGVLVHCLAGISRSVTVTVAY 307
Query 435 LCFCVGLDVRKVNFSVS 451
L ++K+N S++
Sbjct 308 L-------MQKLNLSMN 317
> dre:568887 zgc:172281 (EC:3.1.3.16); K14165 dual specificity
phosphatase [EC:3.1.3.16 3.1.3.48]
Length=189
Score = 36.2 bits (82), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 37/85 (43%), Gaps = 2/85 (2%)
Query 373 QLYESAGLRYVWMPTEDMSD-ACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAV 431
+ Y + Y + +D D A A I AL +NG V+VHC GV RS V
Sbjct 76 RFYRDMNIDYYGVEADDSFDFAISGFFYATARFIRAALSKNG-RVFVHCLMGVSRSATLV 134
Query 432 GAYLCFCVGLDVRKVNFSVSTKRPV 456
A+L C L + + +V R +
Sbjct 135 LAFLMICEDLTLMEAIKAVRQHRDI 159
> dre:100007304 dual specificity phosphatase 10-like
Length=904
Score = 36.2 bits (82), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 35/144 (24%), Positives = 59/144 (40%), Gaps = 24/144 (16%)
Query 315 SRILPNLYVGSCPRQLKHIRQLKEELKVTVVFNMQTADDMKTNFPDPLASKRTPEAAGQL 374
S ILP L++G+ + ++L + + N+ ++ T+ P L
Sbjct 745 SPILPFLFLGN--------ERDAQDLDLLLHLNIGFVVNVTTHLP--------------L 782
Query 375 Y--ESAGLRYVWMPTEDMSDACRKLAVAQCALILDALIQNGHSVYVHCNAGVGRSVAAVG 432
Y ++ +RY +P D S + + ++ Q G V VHC AGV RS V
Sbjct 783 YHLDTGLVRYKRLPATDNSKQNLRQYFEEVFEFIEEAHQCGRGVLVHCQAGVSRSATIVI 842
Query 433 AYLCFCVGLDVRKVNFSVSTKRPV 456
AYL + + V +RP+
Sbjct 843 AYLMKHTLMTMTDAYKYVRGRRPI 866
Lambda K H
0.319 0.134 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 21416549680
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40