bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3075_orf2
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
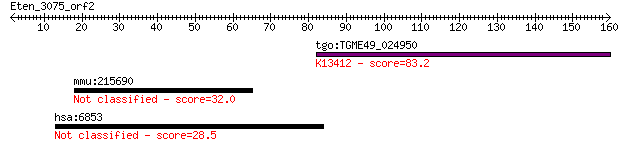
tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.1... 83.2 4e-16
mmu:215690 Nav1, 9530089B19, 9930003A20Rik, C230080M11Rik, Pom... 32.0 0.88
hsa:6853 SYN1, SYN1a, SYN1b, SYNI; synapsin I 28.5 9.1
> tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=681
Score = 83.2 bits (204), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 47/105 (44%), Positives = 57/105 (54%), Gaps = 27/105 (25%)
Query 82 ADHHCHGPE-----EKPHFFSLCTKCATDGKDKPL---REPAKGFITRSESYVNASAASR 133
AD+ C G + KPHFFSLCTKCA +GKDK REP +G I RSES++NAS ASR
Sbjct 92 ADYGCRGGDCCDTSTKPHFFSLCTKCANEGKDKSQHDRREPVRGLIIRSESFMNASEASR 151
Query 134 WEVDELC-------------------GKEPHKEPSGPVFDRSRFV 159
WE++ C G +P PVFDRS+ V
Sbjct 152 WEIEAQCNSTCPGDGFDGASGTTAADGIPASPQPEAPVFDRSKIV 196
> mmu:215690 Nav1, 9530089B19, 9930003A20Rik, C230080M11Rik, Pomfil3,
Unc53h1, mKIAA1151, steerin-1; neuron navigator 1
Length=1875
Score = 32.0 bits (71), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 26/54 (48%), Gaps = 7/54 (12%)
Query 18 DGEGAEPRPAAAAAAAAAAAAAAAAAAAAAGGG-------GGPPASNLQKDKRL 64
DG G P+ A AA + + A A+AA G GG P SNL+K K L
Sbjct 41 DGRGMLPKRAKAAGGSGSMAKASAAELKVFKSGSVDSRVPGGLPTSNLRKQKSL 94
> hsa:6853 SYN1, SYN1a, SYN1b, SYNI; synapsin I
Length=705
Score = 28.5 bits (62), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 30/71 (42%), Gaps = 7/71 (9%)
Query 13 RPDSPDGEGAEPRPAAAAAAAAAAAAAAAAAAAAAGGGGGPPASNLQKDKRLDTAEGRPL 72
RP P G P+P A + A AAA GGPP L K + L A P
Sbjct 622 RPSGP-GPAGRPKPQLAQKPSQDVPPPATAAA------GGPPHPQLNKSQSLTNAFNLPE 674
Query 73 ESPCRPNCAAD 83
+P RP+ + D
Sbjct 675 PAPPRPSLSQD 685
Lambda K H
0.311 0.128 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3647184800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40