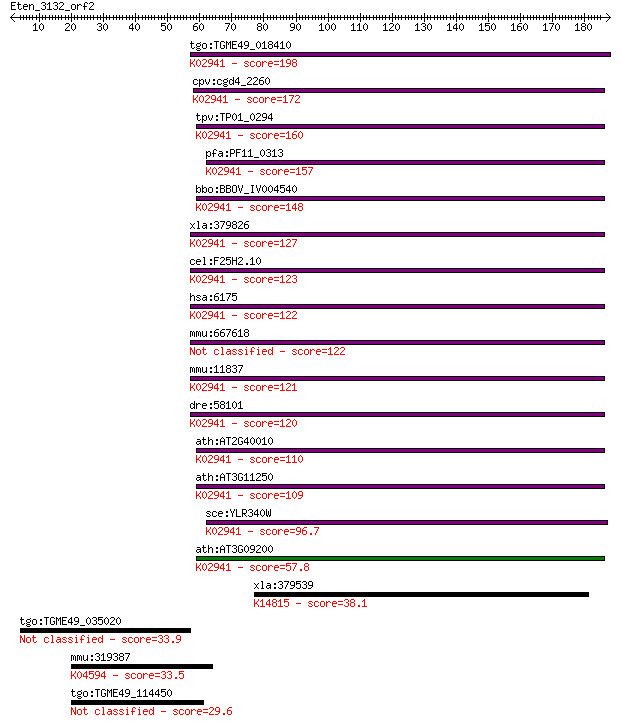
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3132_orf2
Length=187
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_018410 60S acidic ribosomal protein P0 (EC:4.2.99.1... 198 1e-50
cpv:cgd4_2260 ribosomal protein PO like protein of the L10 fam... 172 4e-43
tpv:TP01_0294 60S acidic ribosomal protein, P0; K02941 large s... 160 2e-39
pfa:PF11_0313 60S ribosomal protein P0; K02941 large subunit r... 157 2e-38
bbo:BBOV_IV004540 23.m05776; phosphoriboprotein P0; K02941 lar... 148 1e-35
xla:379826 rplp0, MGC53408, arbp; ribosomal protein, large, P0... 127 2e-29
cel:F25H2.10 rpa-0; Replication Protein A homolog family membe... 123 3e-28
hsa:6175 RPLP0, L10E, MGC111226, MGC88175, P0, PRLP0, RPP0; ri... 122 1e-27
mmu:667618 Gm8730, EG667618; predicted pseudogene 8730 122 1e-27
mmu:11837 Rplp0, 36B4, Arbp, MGC107165, MGC107166; ribosomal p... 121 1e-27
dre:58101 rplp0, arp, fb04a12, wu:fb15a08, wu:fk48c12; ribosom... 120 2e-27
ath:AT2G40010 60S acidic ribosomal protein P0 (RPP0A); K02941 ... 110 3e-24
ath:AT3G11250 60S acidic ribosomal protein P0 (RPP0C); K02941 ... 109 5e-24
sce:YLR340W RPP0, RPL10E; P0; L10E; A0; K02941 large subunit r... 96.7 4e-20
ath:AT3G09200 60S acidic ribosomal protein P0 (RPP0B); K02941 ... 57.8 2e-08
xla:379539 mrto4, MGC68920, mrt4; mRNA turnover 4 homolog; K14... 38.1 0.020
tgo:TGME49_035020 coatomer protein complex subunit beta, putative 33.9 0.32
mmu:319387 Lphn3, 5430402I23Rik, CIRL-3, D130075K09Rik, Gm1379... 33.5 0.42
tgo:TGME49_114450 hypothetical protein 29.6 5.6
> tgo:TGME49_018410 60S acidic ribosomal protein P0 (EC:4.2.99.18);
K02941 large subunit ribosomal protein LP0
Length=314
Score = 198 bits (503), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 93/131 (70%), Positives = 110/131 (83%), Gaps = 0/131 (0%)
Query 57 KERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRT 116
K ++ KR+ YF RL LL ++PRVLVV ADHVGSKQ+A IR+ALRG+A VLMGKNT IRT
Sbjct 5 KGKSDKRKTYFSRLFALLEKYPRVLVVEADHVGSKQMADIRLALRGKAVVLMGKNTMIRT 64
Query 117 ALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
AL+Q++ + PQL+ LLPLVRLNVGFIFC DPA VR +V ++KVPAPA+QGV AP DVFI
Sbjct 65 ALKQKMSEMPQLEKLLPLVRLNVGFIFCIEDPAEVRRIVAENKVPAPARQGVFAPIDVFI 124
Query 177 PAGPTGMDPGS 187
PAGPTGMDPGS
Sbjct 125 PAGPTGMDPGS 135
> cpv:cgd4_2260 ribosomal protein PO like protein of the L10 family
; K02941 large subunit ribosomal protein LP0
Length=318
Score = 172 bits (437), Expect = 4e-43, Method: Compositional matrix adjust.
Identities = 76/128 (59%), Positives = 103/128 (80%), Gaps = 0/128 (0%)
Query 58 ERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTA 117
E+AAK+++YF RL + +PR+LV +ADHVGSKQ+A IR+ALRG+A VLMGKNT IRTA
Sbjct 13 EKAAKKKQYFERLSEYATSYPRILVANADHVGSKQMADIRLALRGKAAVLMGKNTMIRTA 72
Query 118 LRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFIP 177
L+Q L P+L+ L+ LVRLNVG IFC +P+ VR ++++++VPAPA+QGV AP +V +P
Sbjct 73 LKQMLGSHPELEKLIELVRLNVGLIFCIDEPSEVRKIIEEYRVPAPARQGVIAPCNVVVP 132
Query 178 AGPTGMDP 185
AG TG+DP
Sbjct 133 AGATGLDP 140
> tpv:TP01_0294 60S acidic ribosomal protein, P0; K02941 large
subunit ribosomal protein LP0
Length=321
Score = 160 bits (406), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 69/127 (54%), Positives = 95/127 (74%), Gaps = 0/127 (0%)
Query 59 RAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTAL 118
++ K++ YF RL L+ + ++L+VS DHVGS+Q+A +R +LRG AT+LMGKNT IRTAL
Sbjct 15 KSEKKKLYFERLTSLMRTYSKILIVSVDHVGSRQMASVRHSLRGMATILMGKNTVIRTAL 74
Query 119 RQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFIPA 178
++ P ++ + V+LN GF+FC ADP VR V+ ++VPAPAKQGV AP+DVFIPA
Sbjct 75 QKNFPDSPDVEKVAQCVKLNTGFVFCEADPMEVREVILNNRVPAPAKQGVIAPSDVFIPA 134
Query 179 GPTGMDP 185
G TG+DP
Sbjct 135 GSTGLDP 141
> pfa:PF11_0313 60S ribosomal protein P0; K02941 large subunit
ribosomal protein LP0
Length=316
Score = 157 bits (398), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 67/124 (54%), Positives = 97/124 (78%), Gaps = 0/124 (0%)
Query 62 KRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTALRQQ 121
K++ Y +L L+ ++ ++L+V D+VGS Q+A +R +LRG+AT+LMGKNT+IRTAL++
Sbjct 9 KKQMYIEKLSSLIQQYSKILIVHVDNVGSNQMASVRKSLRGKATILMGKNTRIRTALKKN 68
Query 122 LQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFIPAGPT 181
LQ PQ++ LLPLV+LN+GF+FC+ D + +R ++ +K PAPA+ GV AP DVFIP GPT
Sbjct 69 LQAVPQIEKLLPLVKLNMGFVFCKDDLSEIRNIILDNKSPAPARLGVIAPIDVFIPPGPT 128
Query 182 GMDP 185
GMDP
Sbjct 129 GMDP 132
> bbo:BBOV_IV004540 23.m05776; phosphoriboprotein P0; K02941 large
subunit ribosomal protein LP0
Length=312
Score = 148 bits (374), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 66/127 (51%), Positives = 88/127 (69%), Gaps = 0/127 (0%)
Query 59 RAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTAL 118
+ K++ YF RL L+ +P++L+VS D+VGS+Q+A +R +LRG+A +L+GKNT IR L
Sbjct 6 KQEKKKAYFERLTHLVKTYPQILIVSVDYVGSRQMAHVRHSLRGKAEILIGKNTMIRMVL 65
Query 119 RQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFIPA 178
+ LL V+LNVGF+FC DP VR ++ +KVPAPAKQGV AP DVFI A
Sbjct 66 NTSFPNSEAISKLLSCVKLNVGFVFCMGDPLEVRRIILDNKVPAPAKQGVIAPCDVFISA 125
Query 179 GPTGMDP 185
G TGMDP
Sbjct 126 GATGMDP 132
> xla:379826 rplp0, MGC53408, arbp; ribosomal protein, large,
P0; K02941 large subunit ribosomal protein LP0
Length=315
Score = 127 bits (320), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 59/129 (45%), Positives = 89/129 (68%), Gaps = 0/129 (0%)
Query 57 KERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRT 116
++RA + YF +++QLL ++P+ +V AD+VGSKQ+ IR++LRG+A VLMGKNT +R
Sbjct 4 EDRATWKSNYFLKIIQLLDDYPKCFIVGADNVGSKQMQQIRMSLRGKAVVLMGKNTMMRK 63
Query 117 ALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
A+R L+ P L+ LL ++ NVGF+F + D VR ++ +KVPA A+ G AP +V +
Sbjct 64 AIRGHLENNPALEKLLSHIKGNVGFVFTKEDLTEVRDMLLANKVPASARAGAIAPCEVTV 123
Query 177 PAGPTGMDP 185
PA TG+ P
Sbjct 124 PAQNTGLGP 132
> cel:F25H2.10 rpa-0; Replication Protein A homolog family member
(rpa-0); K02941 large subunit ribosomal protein LP0
Length=312
Score = 123 bits (309), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 59/129 (45%), Positives = 85/129 (65%), Gaps = 0/129 (0%)
Query 57 KERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRT 116
++R+ + YF +L++L E+P+ L+V D+VGSKQ+ IR A+RG A +LMGKNT IR
Sbjct 4 EDRSTWKANYFTKLVELFEEYPKCLLVGVDNVGSKQMQEIRQAMRGHAEILMGKNTMIRK 63
Query 117 ALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
ALR L + P L+ LLP + NVGF+F + D +R+ + +++ APAK G AP DV +
Sbjct 64 ALRGHLGKNPSLEKLLPHIVENVGFVFTKEDLGEIRSKLLENRKGAPAKAGAIAPCDVKL 123
Query 177 PAGPTGMDP 185
P TGM P
Sbjct 124 PPQNTGMGP 132
> hsa:6175 RPLP0, L10E, MGC111226, MGC88175, P0, PRLP0, RPP0;
ribosomal protein, large, P0; K02941 large subunit ribosomal
protein LP0
Length=317
Score = 122 bits (305), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 60/129 (46%), Positives = 90/129 (69%), Gaps = 0/129 (0%)
Query 57 KERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRT 116
++RA + YF +++QLL ++P+ +V AD+VGSKQ+ IR++LRG+A VLMGKNT +R
Sbjct 4 EDRATWKSNYFLKIIQLLDDYPKCFIVGADNVGSKQMQQIRMSLRGKAVVLMGKNTMMRK 63
Query 117 ALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
A+R L+ P L+ LLP +R NVGF+F + D +R ++ +KVPA A+ G AP +V +
Sbjct 64 AIRGHLENNPALEKLLPHIRGNVGFVFTKEDLTEIRDMLLANKVPAAARAGAIAPCEVTV 123
Query 177 PAGPTGMDP 185
PA TG+ P
Sbjct 124 PAQNTGLGP 132
> mmu:667618 Gm8730, EG667618; predicted pseudogene 8730
Length=317
Score = 122 bits (305), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 60/129 (46%), Positives = 90/129 (69%), Gaps = 0/129 (0%)
Query 57 KERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRT 116
++RA + YF +++QLL ++P+ +V AD+VGSKQ+ IR++LRG+A VLMGKNT +R
Sbjct 4 EDRATWKSNYFLKIIQLLDDYPKCFIVGADNVGSKQMQQIRMSLRGKAVVLMGKNTMMRK 63
Query 117 ALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
A+R L+ P L+ LLP +R NVGF+F + D +R ++ +KVPA A+ G AP +V +
Sbjct 64 AIRGHLENNPALEKLLPHIRGNVGFVFTKEDLTEIRDMLLANKVPAAARAGAIAPCEVTV 123
Query 177 PAGPTGMDP 185
PA TG+ P
Sbjct 124 PAQNTGLGP 132
> mmu:11837 Rplp0, 36B4, Arbp, MGC107165, MGC107166; ribosomal
protein, large, P0; K02941 large subunit ribosomal protein
LP0
Length=317
Score = 121 bits (304), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 60/129 (46%), Positives = 90/129 (69%), Gaps = 0/129 (0%)
Query 57 KERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRT 116
++RA + YF +++QLL ++P+ +V AD+VGSKQ+ IR++LRG+A VLMGKNT +R
Sbjct 4 EDRATWKSNYFLKIIQLLDDYPKCFIVGADNVGSKQMQQIRMSLRGKAVVLMGKNTMMRK 63
Query 117 ALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
A+R L+ P L+ LLP +R NVGF+F + D +R ++ +KVPA A+ G AP +V +
Sbjct 64 AIRGHLENNPALEKLLPHIRGNVGFVFTKEDLTEIRDMLLANKVPAAARAGAIAPCEVTV 123
Query 177 PAGPTGMDP 185
PA TG+ P
Sbjct 124 PAQNTGLGP 132
> dre:58101 rplp0, arp, fb04a12, wu:fb15a08, wu:fk48c12; ribosomal
protein, large, P0; K02941 large subunit ribosomal protein
LP0
Length=319
Score = 120 bits (302), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 61/129 (47%), Positives = 89/129 (68%), Gaps = 0/129 (0%)
Query 57 KERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRT 116
++RA + YF +++QLL + P+ +V AD+VGSKQ+ IR++LRG+A VLMGKNT +R
Sbjct 4 EDRATWKSNYFLKIIQLLDDFPKCFIVGADNVGSKQMQTIRLSLRGKAVVLMGKNTMMRK 63
Query 117 ALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
A+R L+ P L+ LLP +R NVGF+F + D VR ++ +KVPA A+ G AP +V +
Sbjct 64 AIRGHLENNPALERLLPHIRGNVGFVFTKEDLTEVRDLLLANKVPAAARAGAIAPCEVTV 123
Query 177 PAGPTGMDP 185
PA TG+ P
Sbjct 124 PAQNTGLGP 132
> ath:AT2G40010 60S acidic ribosomal protein P0 (RPP0A); K02941
large subunit ribosomal protein LP0
Length=317
Score = 110 bits (275), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 58/129 (44%), Positives = 82/129 (63%), Gaps = 2/129 (1%)
Query 59 RAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTAL 118
+A K+ Y +L QLL E+ ++LVV+AD+VGS QL IR LRG + VLMGKNT ++ ++
Sbjct 7 KAEKKIVYDSKLCQLLNEYSQILVVAADNVGSTQLQNIRKGLRGDSVVLMGKNTMMKRSV 66
Query 119 RQQLQQQPQ--LQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
R + +LLPL++ NVG IF + D V V ++KV APA+ G+ AP DV +
Sbjct 67 RIHADKTGNQAFLSLLPLLQGNVGLIFTKGDLKEVSEEVAKYKVGAPARVGLVAPIDVVV 126
Query 177 PAGPTGMDP 185
G TG+DP
Sbjct 127 QPGNTGLDP 135
> ath:AT3G11250 60S acidic ribosomal protein P0 (RPP0C); K02941
large subunit ribosomal protein LP0
Length=323
Score = 109 bits (273), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 57/129 (44%), Positives = 82/129 (63%), Gaps = 2/129 (1%)
Query 59 RAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTAL 118
+A K+ Y +L QL+ E+ ++LVV+AD+VGS QL IR LRG + VLMGKNT ++ ++
Sbjct 6 KAEKKIAYDTKLCQLIDEYTQILVVAADNVGSTQLQNIRKGLRGDSVVLMGKNTMMKRSV 65
Query 119 RQQLQQ--QPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
R + + LLPL++ NVG IF + D V V ++KV APA+ G+ AP DV +
Sbjct 66 RIHSENSGNTAILNLLPLLQGNVGLIFTKGDLKEVSEEVAKYKVGAPARVGLVAPIDVVV 125
Query 177 PAGPTGMDP 185
G TG+DP
Sbjct 126 QPGNTGLDP 134
> sce:YLR340W RPP0, RPL10E; P0; L10E; A0; K02941 large subunit
ribosomal protein LP0
Length=312
Score = 96.7 bits (239), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 54/125 (43%), Positives = 79/125 (63%), Gaps = 0/125 (0%)
Query 62 KRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTALRQQ 121
K+ EYF +L + L E+ + VV D+V S+Q+ +R LRG+A VLMGKNT +R A+R
Sbjct 7 KKAEYFAKLREYLEEYKSLFVVGVDNVSSQQMHEVRKELRGRAVVLMGKNTMVRRAIRGF 66
Query 122 LQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFIPAGPT 181
L P + LLP V+ NVGF+F ++ V+ ++V APA+ G AP D+++ A T
Sbjct 67 LSDLPDFEKLLPFVKGNVGFVFTNEPLTEIKNVIVSNRVAAPARAGAVAPEDIWVRAVNT 126
Query 182 GMDPG 186
GM+PG
Sbjct 127 GMEPG 131
> ath:AT3G09200 60S acidic ribosomal protein P0 (RPP0B); K02941
large subunit ribosomal protein LP0
Length=287
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 42/127 (33%), Positives = 60/127 (47%), Gaps = 31/127 (24%)
Query 59 RAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTAL 118
+A K+ Y +L QL+ E+ ++LV S V + + T NT I
Sbjct 6 KAEKKIAYDTKLCQLIDEYTQILVRS-------------VRIHSENT----GNTAILN-- 46
Query 119 RQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFIPA 178
LLPL++ NVG IF + D V V ++KV APA+ G+ AP DV +
Sbjct 47 ------------LLPLLQGNVGLIFTKGDLKEVSEEVAKYKVGAPARVGLVAPIDVVVQP 94
Query 179 GPTGMDP 185
G TG+DP
Sbjct 95 GNTGLDP 101
> xla:379539 mrto4, MGC68920, mrt4; mRNA turnover 4 homolog; K14815
mRNA turnover protein 4
Length=240
Score = 38.1 bits (87), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 24/106 (22%), Positives = 48/106 (45%), Gaps = 3/106 (2%)
Query 77 HPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTALRQQL--QQQPQLQALLPL 134
+ + V+S +++ + +L +R A + + + GKN + A+ + + + + L L
Sbjct 37 YKYIFVLSVENMRNNKLKDVRNAWK-HSRLFFGKNKVMMVAMGKGVSDEYKDNLHKLSKC 95
Query 135 VRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFIPAGP 180
++ VG +F V Q+K A+ G A V + AGP
Sbjct 96 LKGEVGLLFTNRTEKEVEEWFDQYKETDFARAGNKATYSVVLDAGP 141
> tgo:TGME49_035020 coatomer protein complex subunit beta, putative
Length=1256
Score = 33.9 bits (76), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 21/53 (39%), Positives = 32/53 (60%), Gaps = 4/53 (7%)
Query 4 FWRSLPGIPSDLWLKRLPGRRLCQSPQAPESSAHASAGAASGFRASAAAGAMG 56
F +L G+P+D+ RLP SP APE++ AS+ A +R++ A GA+G
Sbjct 981 FDETLSGVPADM--HRLPHSHAASSPVAPEAA--ASSSQAPLYRSAVAEGALG 1029
> mmu:319387 Lphn3, 5430402I23Rik, CIRL-3, D130075K09Rik, Gm1379,
LEC3, MGC99439, mKIAA0768; latrophilin 3; K04594 latrophilin
3
Length=1543
Score = 33.5 bits (75), Expect = 0.42, Method: Composition-based stats.
Identities = 20/47 (42%), Positives = 22/47 (46%), Gaps = 3/47 (6%)
Query 20 LPGRRLCQSPQAPESSAH---ASAGAASGFRASAAAGAMGKERAAKR 63
LP R L Q P A S+AH GAA G R A GA +A R
Sbjct 44 LPPRHLLQQPAAERSTAHRGQGPRGAARGVRGPGAPGAQIAAQAFSR 90
> tgo:TGME49_114450 hypothetical protein
Length=331
Score = 29.6 bits (65), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 22/46 (47%), Positives = 27/46 (58%), Gaps = 6/46 (13%)
Query 20 LPGRRLCQSP-----QAPESSAHASAGAASGFRASAAAGAMGKERA 60
LP +RLC SP Q P SSAHA+ G+AS RA+A G+ A
Sbjct 18 LPQQRLCSSPGFGPRQTP-SSAHAAMGSASLSRAAAHHGSYSSSMA 62
Lambda K H
0.321 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5235419772
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40