bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3150_orf1
Length=169
Score E
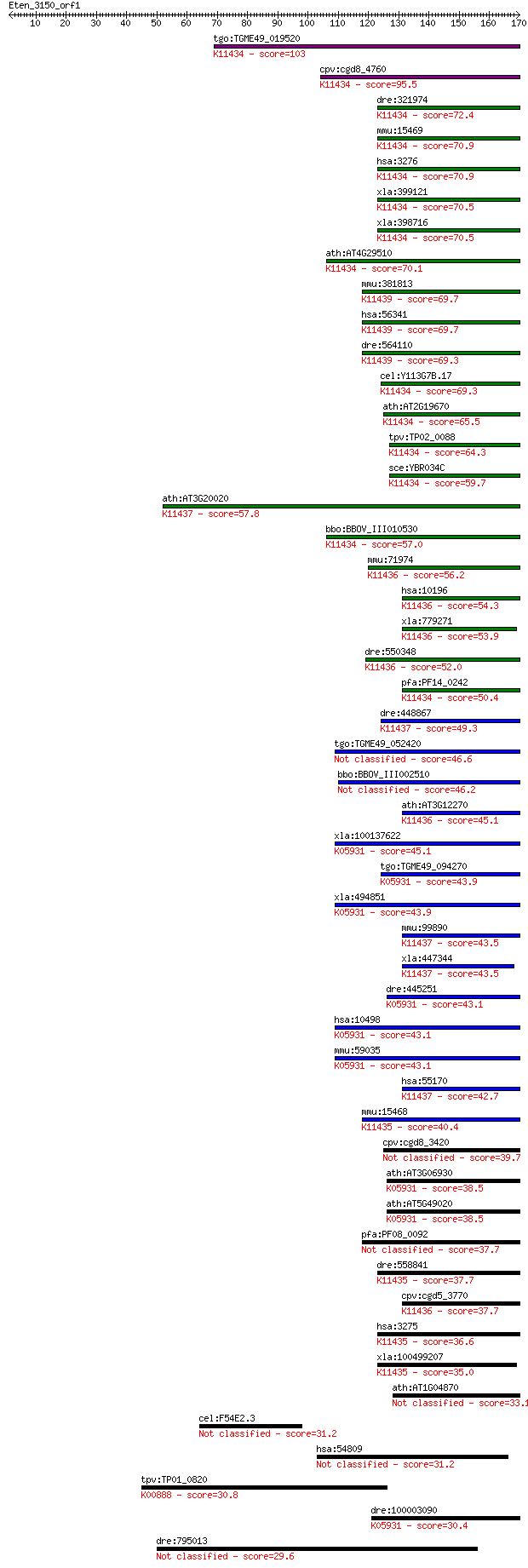
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_019520 arginine N-methyltransferase 1 (EC:2.1.1.125... 103 3e-22
cpv:cgd8_4760 arginine n-methyltransferase ; K11434 protein ar... 95.5 7e-20
dre:321974 prmt1, MGC66201, fb39h07, hrmt1l2, wu:fb39h07, zf1,... 72.4 7e-13
mmu:15469 Prmt1, 6720434D09Rik, AW214366, Hrmt1l2, Mrmt1; prot... 70.9 2e-12
hsa:3276 PRMT1, ANM1, HCP1, HRMT1L2, IR1B4; protein arginine m... 70.9 2e-12
xla:399121 prmt1-a, Hrmt1l2, anm1, hcp1, ir1b4, prmt1, xPRMT1;... 70.5 2e-12
xla:398716 prmt1-b, MGC130744, anm1, hcp1, hrmt1l2, ir1b4, prm... 70.5 3e-12
ath:AT4G29510 PRMT11; PRMT11 (ARGININE METHYLTRANSFERASE 11); ... 70.1 3e-12
mmu:381813 Prmt8, Hrmt1l3, Hrmt1l4; protein arginine N-methylt... 69.7 4e-12
hsa:56341 PRMT8, HRMT1L3, HRMT1L4; protein arginine methyltran... 69.7 4e-12
dre:564110 prmt8b, HRMT1L3, fj34f03, hrmt1l4, prmt8, wu:fj34f0... 69.3 6e-12
cel:Y113G7B.17 hypothetical protein; K11434 protein arginine N... 69.3 6e-12
ath:AT2G19670 PRMT1A; PRMT1A (PROTEIN ARGININE METHYLTRANSFERA... 65.5 8e-11
tpv:TP02_0088 arginine N-methyltransferase; K11434 protein arg... 64.3 2e-10
sce:YBR034C HMT1, HCP1, ODP1, RMT1; Hmt1p (EC:2.1.1.-); K11434... 59.7 4e-09
ath:AT3G20020 PRMT6; PRMT6 (PROTEIN ARGININE METHYLTRANSFERASE... 57.8 2e-08
bbo:BBOV_III010530 17.m07911; hypothetical protein; K11434 pro... 57.0 3e-08
mmu:71974 Prmt3, 2010005E20Rik, 2410018A17Rik, AL033309, Hrmt1... 56.2 5e-08
hsa:10196 PRMT3, HRMT1L3; protein arginine methyltransferase 3... 54.3 2e-07
xla:779271 prmt3, MGC154481; protein arginine methyltransferas... 53.9 2e-07
dre:550348 prmt3, hrmt1l3, zgc:112498; protein arginine methyl... 52.0 1e-06
pfa:PF14_0242 arginine-N-methyltransferase, putative; K11434 p... 50.4 3e-06
dre:448867 prmt6, hrmt1l6, hrmt6, im:6908706; protein arginine... 49.3 6e-06
tgo:TGME49_052420 hypothetical protein 46.6 4e-05
bbo:BBOV_III002510 17.m07242; hypothetical protein 46.2 5e-05
ath:AT3G12270 PRMT3; PRMT3 (PROTEIN ARGININE METHYLTRANSFERASE... 45.1 1e-04
xla:100137622 coactivator-associated arginine methyltransferas... 45.1 1e-04
tgo:TGME49_094270 arginine methyltransferase protein, putative... 43.9 2e-04
xla:494851 carm1; coactivator-associated arginine methyltransf... 43.9 3e-04
mmu:99890 Prmt6, AW124876, BB233495, Hrmt1l6; protein arginine... 43.5 3e-04
xla:447344 prmt6, MGC83989; protein arginine methyltransferase... 43.5 4e-04
dre:445251 carm1, PRMT4, si:dkey-204f11.63, zgc:100805; coacti... 43.1 4e-04
hsa:10498 CARM1, PRMT4; coactivator-associated arginine methyl... 43.1 4e-04
mmu:59035 Carm1, MGC46828, Prmt4; coactivator-associated argin... 43.1 4e-04
hsa:55170 PRMT6, FLJ10559, FLJ51477, HRMT1L6; protein arginine... 42.7 5e-04
mmu:15468 Prmt2, AI504737, Hrmt1l1, MGC148245, MGC148246; prot... 40.4 0.003
cpv:cgd8_3420 hypothetical protein 39.7 0.005
ath:AT3G06930 PRMT4B; protein arginine N-methyltransferase fam... 38.5 0.011
ath:AT5G49020 PRMT4A; PRMT4A (PROTEIN ARGININE METHYLTRANSFERA... 38.5 0.012
pfa:PF08_0092 methyltransferase, putative 37.7 0.017
dre:558841 prmt2, MGC158247, hrmt1l1, si:ch211-226c11.1, zgc:1... 37.7 0.018
cpv:cgd5_3770 arginine N-methyltransferase ; K11436 protein ar... 37.7 0.019
hsa:3275 PRMT2, HRMT1L1, MGC111373; protein arginine methyltra... 36.6 0.039
xla:100499207 prmt2; protein arginine methyltransferase 2 (EC:... 35.0 0.11
ath:AT1G04870 PRMT10; PRMT10; [myelin basic protein]-arginine ... 33.1 0.42
cel:F54E2.3 ketn-1; KETtiN (Drosophila actin-binding) homolog ... 31.2 2.0
hsa:54809 SAMD9, C7orf5, DRIF1, FLJ20073, KIAA2004, NFTC, OEF1... 31.2 2.0
tpv:TP01_0820 phosphatidylinositol 4-kinase; K00888 phosphatid... 30.8 2.0
dre:100003090 carm1l; coactivator-associated arginine methyltr... 30.4 2.8
dre:795013 phospholipase A2 receptor 1-like 29.6 4.8
> tgo:TGME49_019520 arginine N-methyltransferase 1 (EC:2.1.1.125);
K11434 protein arginine N-methyltransferase 1 [EC:2.1.1.-]
Length=392
Score = 103 bits (257), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 54/101 (53%), Positives = 64/101 (63%), Gaps = 6/101 (5%)
Query 69 GNGISANNGGSQTGNGSVRGGRGGRPAYAQSLQCQKLSPEEKAAFSADWKQLQQDDISSA 128
G + G + +G RG G A L C + + KAAF+ DWK L+Q+++SSA
Sbjct 16 GRVVVPTGGSRRENSGPARGPAGFS---ATVLPCDEAT---KAAFAKDWKDLEQENLSSA 69
Query 129 DYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
DYYFNSYAHF IHEEMIKDSVRT YQRAI N HLF KV
Sbjct 70 DYYFNSYAHFGIHEEMIKDSVRTGCYQRAICQNAHLFANKV 110
> cpv:cgd8_4760 arginine n-methyltransferase ; K11434 protein
arginine N-methyltransferase 1 [EC:2.1.1.-]
Length=348
Score = 95.5 bits (236), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 44/66 (66%), Positives = 51/66 (77%), Gaps = 0/66 (0%)
Query 104 KLSPEEKAAFSADWKQLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQH 163
K + EE F W L Q+DISSADYYFNSYAHF IHEEM+KDSVRT SYQ+AIM N+H
Sbjct 3 KPTEEELKEFKNSWNPLVQEDISSADYYFNSYAHFGIHEEMLKDSVRTGSYQKAIMSNKH 62
Query 164 LFRGKV 169
LF+ K+
Sbjct 63 LFQDKI 68
> dre:321974 prmt1, MGC66201, fb39h07, hrmt1l2, wu:fb39h07, zf1,
zgc:66201; protein arginine methyltransferase 1 (EC:2.1.1.125);
K11434 protein arginine N-methyltransferase 1 [EC:2.1.1.-]
Length=341
Score = 72.4 bits (176), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 29/47 (61%), Positives = 39/47 (82%), Gaps = 0/47 (0%)
Query 123 DDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
+D++S DYYF+SYAHF IHEEM+KD VRT +Y+ ++ N+HLFR KV
Sbjct 15 EDMTSKDYYFDSYAHFGIHEEMLKDEVRTLTYRNSMFHNKHLFRDKV 61
> mmu:15469 Prmt1, 6720434D09Rik, AW214366, Hrmt1l2, Mrmt1; protein
arginine N-methyltransferase 1 (EC:2.1.1.125); K11434
protein arginine N-methyltransferase 1 [EC:2.1.1.-]
Length=371
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 28/47 (59%), Positives = 39/47 (82%), Gaps = 0/47 (0%)
Query 123 DDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
+D++S DYYF+SYAHF IHEEM+KD VRT +Y+ ++ N+HLF+ KV
Sbjct 45 EDMTSKDYYFDSYAHFGIHEEMLKDEVRTLTYRNSMFHNRHLFKDKV 91
> hsa:3276 PRMT1, ANM1, HCP1, HRMT1L2, IR1B4; protein arginine
methyltransferase 1 (EC:2.1.1.125); K11434 protein arginine
N-methyltransferase 1 [EC:2.1.1.-]
Length=371
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 28/47 (59%), Positives = 39/47 (82%), Gaps = 0/47 (0%)
Query 123 DDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
+D++S DYYF+SYAHF IHEEM+KD VRT +Y+ ++ N+HLF+ KV
Sbjct 45 EDMTSKDYYFDSYAHFGIHEEMLKDEVRTLTYRNSMFHNRHLFKDKV 91
> xla:399121 prmt1-a, Hrmt1l2, anm1, hcp1, ir1b4, prmt1, xPRMT1;
protein arginine methyltransferase 1 (EC:2.1.1.125); K11434
protein arginine N-methyltransferase 1 [EC:2.1.1.-]
Length=369
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 28/47 (59%), Positives = 39/47 (82%), Gaps = 0/47 (0%)
Query 123 DDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
+D++S DYYF+SYAHF IHEEM+KD VRT +Y+ ++ N+HLF+ KV
Sbjct 43 EDMTSKDYYFDSYAHFGIHEEMLKDEVRTLTYRNSMFHNRHLFKDKV 89
> xla:398716 prmt1-b, MGC130744, anm1, hcp1, hrmt1l2, ir1b4, prmt1,
prmt1b, xPRMT1b, xprmt1; protein arginine methyltransferase
1 (EC:2.1.1.125); K11434 protein arginine N-methyltransferase
1 [EC:2.1.1.-]
Length=351
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 28/47 (59%), Positives = 39/47 (82%), Gaps = 0/47 (0%)
Query 123 DDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
+D++S DYYF+SYAHF IHEEM+KD VRT +Y+ ++ N+HLF+ KV
Sbjct 25 EDMTSKDYYFDSYAHFGIHEEMLKDEVRTLTYRNSMFHNRHLFKDKV 71
> ath:AT4G29510 PRMT11; PRMT11 (ARGININE METHYLTRANSFERASE 11);
protein-arginine N-methyltransferase; K11434 protein arginine
N-methyltransferase 1 [EC:2.1.1.-]
Length=390
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 33/68 (48%), Positives = 48/68 (70%), Gaps = 5/68 (7%)
Query 106 SPEEKAAF----SADWKQLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDN 161
+P++++ F SAD ++ DD +SADYYF+SY+HF IHEEM+KD VRT +YQ I N
Sbjct 44 TPQDESMFDAGESADTAEVT-DDTTSADYYFDSYSHFGIHEEMLKDVVRTKTYQNVIYQN 102
Query 162 QHLFRGKV 169
+ L + K+
Sbjct 103 KFLIKDKI 110
> mmu:381813 Prmt8, Hrmt1l3, Hrmt1l4; protein arginine N-methyltransferase
8; K11439 protein arginine N-methyltransferase
8 [EC:2.1.1.-]
Length=379
Score = 69.7 bits (169), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 28/52 (53%), Positives = 41/52 (78%), Gaps = 0/52 (0%)
Query 118 KQLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
K L ++++S DYYF+SYAHF IHEEM+KD VRT +Y+ ++ N+H+F+ KV
Sbjct 48 KLLNPEEMTSRDYYFDSYAHFGIHEEMLKDEVRTLTYRNSMYHNKHVFKDKV 99
> hsa:56341 PRMT8, HRMT1L3, HRMT1L4; protein arginine methyltransferase
8 (EC:2.1.1.-); K11439 protein arginine N-methyltransferase
8 [EC:2.1.1.-]
Length=394
Score = 69.7 bits (169), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 28/52 (53%), Positives = 41/52 (78%), Gaps = 0/52 (0%)
Query 118 KQLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
K L ++++S DYYF+SYAHF IHEEM+KD VRT +Y+ ++ N+H+F+ KV
Sbjct 63 KLLNPEEMTSRDYYFDSYAHFGIHEEMLKDEVRTLTYRNSMYHNKHVFKDKV 114
> dre:564110 prmt8b, HRMT1L3, fj34f03, hrmt1l4, prmt8, wu:fj34f03,
zfL3; protein arginine methyltransferase 8b (EC:2.1.1.-);
K11439 protein arginine N-methyltransferase 8 [EC:2.1.1.-]
Length=334
Score = 69.3 bits (168), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 27/52 (51%), Positives = 41/52 (78%), Gaps = 0/52 (0%)
Query 118 KQLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
K L ++++S DYYF+SYAHF IHEEM+KD VRT +Y+ ++ N+H+F+ K+
Sbjct 3 KLLNPEEMTSRDYYFDSYAHFGIHEEMLKDEVRTLTYRNSMYHNKHIFKDKI 54
> cel:Y113G7B.17 hypothetical protein; K11434 protein arginine
N-methyltransferase 1 [EC:2.1.1.-]
Length=348
Score = 69.3 bits (168), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 28/46 (60%), Positives = 37/46 (80%), Gaps = 0/46 (0%)
Query 124 DISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
+++S DYYF+SYAHF IHEEM+KD VRT +Y+ +I N HLF+ KV
Sbjct 20 ELTSKDYYFDSYAHFGIHEEMLKDEVRTTTYRNSIYHNSHLFKDKV 65
> ath:AT2G19670 PRMT1A; PRMT1A (PROTEIN ARGININE METHYLTRANSFERASE
1A); protein-arginine N-methyltransferase; K11434 protein
arginine N-methyltransferase 1 [EC:2.1.1.-]
Length=366
Score = 65.5 bits (158), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 28/45 (62%), Positives = 35/45 (77%), Gaps = 0/45 (0%)
Query 125 ISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
I+SADYYF+SY+HF IHEEM+KD VRT SYQ I N+ L + K+
Sbjct 42 ITSADYYFDSYSHFGIHEEMLKDVVRTKSYQDVIYKNKFLIKDKI 86
> tpv:TP02_0088 arginine N-methyltransferase; K11434 protein arginine
N-methyltransferase 1 [EC:2.1.1.-]
Length=373
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 28/43 (65%), Positives = 33/43 (76%), Gaps = 0/43 (0%)
Query 127 SADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
+ D YFNSY++ IHEEM+KDSVRT Y + IM NQHLFR KV
Sbjct 50 NTDVYFNSYSYIGIHEEMLKDSVRTGIYYKTIMTNQHLFRDKV 92
> sce:YBR034C HMT1, HCP1, ODP1, RMT1; Hmt1p (EC:2.1.1.-); K11434
protein arginine N-methyltransferase 1 [EC:2.1.1.-]
Length=348
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 24/43 (55%), Positives = 34/43 (79%), Gaps = 0/43 (0%)
Query 127 SADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
S +YFNSY H+ IHEEM++D+VRT SY+ AI+ N+ LF+ K+
Sbjct 19 SEQHYFNSYDHYGIHEEMLQDTVRTLSYRNAIIQNKDLFKDKI 61
> ath:AT3G20020 PRMT6; PRMT6 (PROTEIN ARGININE METHYLTRANSFERASE
6); methyltransferase; K11437 protein arginine N-methyltransferase
6 [EC:2.1.1.-]
Length=413
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 42/123 (34%), Positives = 57/123 (46%), Gaps = 7/123 (5%)
Query 52 MESGQNHQKGALPSHAKGNGISANNGGSQTGNGSVR-----GGRGGRPAYAQSLQCQKLS 106
M+SG + G H + + G S + G + G R R A L+
Sbjct 1 MQSGGDFSNGFHGDHHRELELEDKQGPSLSSFGRAKKRSHAGARDPRGGLANVLRVSDQL 60
Query 107 PEEKAAFSADWKQLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFR 166
E K+ +++ D A YF+SYAH IHEEMIKD RT +Y+ AIM +Q L
Sbjct 61 GEHKSLETSESSPPPCTDFDVA--YFHSYAHVGIHEEMIKDRARTETYREAIMQHQSLIE 118
Query 167 GKV 169
GKV
Sbjct 119 GKV 121
> bbo:BBOV_III010530 17.m07911; hypothetical protein; K11434 protein
arginine N-methyltransferase 1 [EC:2.1.1.-]
Length=368
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 31/76 (40%), Positives = 43/76 (56%), Gaps = 12/76 (15%)
Query 106 SPEEKAAFSADWKQL------QQDDISSA------DYYFNSYAHFSIHEEMIKDSVRTCS 153
S + ++FS DW + I+ A D YF+SY + IHEEM+KD+VRT +
Sbjct 13 SANQISSFSNDWADFGSNAESLESSINDAEPSGAQDSYFHSYGYIGIHEEMLKDAVRTGT 72
Query 154 YQRAIMDNQHLFRGKV 169
Y +AI N+HLF KV
Sbjct 73 YHKAITQNRHLFENKV 88
> mmu:71974 Prmt3, 2010005E20Rik, 2410018A17Rik, AL033309, Hrmt1l3;
protein arginine N-methyltransferase 3; K11436 protein
arginine N-methyltransferase 3 [EC:2.1.1.-]
Length=528
Score = 56.2 bits (134), Expect = 5e-08, Method: Composition-based stats.
Identities = 26/50 (52%), Positives = 34/50 (68%), Gaps = 3/50 (6%)
Query 120 LQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
LQ+D+ YF+SY H+ IHEEM+KD VRT SY+ I N H+F+ KV
Sbjct 209 LQEDE---DGVYFSSYGHYGIHEEMLKDKVRTESYRDFIYQNPHIFKDKV 255
> hsa:10196 PRMT3, HRMT1L3; protein arginine methyltransferase
3; K11436 protein arginine N-methyltransferase 3 [EC:2.1.1.-]
Length=469
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 22/39 (56%), Positives = 29/39 (74%), Gaps = 0/39 (0%)
Query 131 YFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
YF+SY H+ IHEEM+KD +RT SY+ I N H+F+ KV
Sbjct 158 YFSSYGHYGIHEEMLKDKIRTESYRDFIYQNPHIFKDKV 196
> xla:779271 prmt3, MGC154481; protein arginine methyltransferase
3; K11436 protein arginine N-methyltransferase 3 [EC:2.1.1.-]
Length=519
Score = 53.9 bits (128), Expect = 2e-07, Method: Composition-based stats.
Identities = 22/38 (57%), Positives = 29/38 (76%), Gaps = 0/38 (0%)
Query 131 YFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGK 168
YF+SY HF IHE+M+KD+VRT SY+ I N H+F+ K
Sbjct 208 YFSSYGHFGIHEDMLKDTVRTESYRDFIYQNPHIFKDK 245
> dre:550348 prmt3, hrmt1l3, zgc:112498; protein arginine methyltransferase
3; K11436 protein arginine N-methyltransferase
3 [EC:2.1.1.-]
Length=512
Score = 52.0 bits (123), Expect = 1e-06, Method: Composition-based stats.
Identities = 24/51 (47%), Positives = 36/51 (70%), Gaps = 3/51 (5%)
Query 119 QLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
+L++D+ + YF+SY H+SIHEEM+KD VRT SY+ + N +F+ KV
Sbjct 192 ELREDE---DEAYFSSYGHYSIHEEMLKDKVRTESYRDFMYRNMDVFKDKV 239
> pfa:PF14_0242 arginine-N-methyltransferase, putative; K11434
protein arginine N-methyltransferase 1 [EC:2.1.1.-]
Length=401
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 21/39 (53%), Positives = 28/39 (71%), Gaps = 0/39 (0%)
Query 131 YFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
YFNSY + IHE+MIKD VRT +Y +I N+HL + K+
Sbjct 83 YFNSYNYIHIHEDMIKDEVRTRTYYDSIRKNEHLIKDKI 121
> dre:448867 prmt6, hrmt1l6, hrmt6, im:6908706; protein arginine
methyltransferase 6 (EC:2.1.1.125); K11437 protein arginine
N-methyltransferase 6 [EC:2.1.1.-]
Length=355
Score = 49.3 bits (116), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 24/47 (51%), Positives = 31/47 (65%), Gaps = 1/47 (2%)
Query 124 DISSADY-YFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
D S+ DY YF+SY+ +IHEEMI D+VRT +Y+ I N GKV
Sbjct 18 DRSTEDYMYFDSYSDVTIHEEMIADTVRTNTYRMGIFKNSKSIEGKV 64
> tgo:TGME49_052420 hypothetical protein
Length=802
Score = 46.6 bits (109), Expect = 4e-05, Method: Composition-based stats.
Identities = 25/61 (40%), Positives = 32/61 (52%), Gaps = 0/61 (0%)
Query 109 EKAAFSADWKQLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGK 168
E A D Q Q++ YF YA SIH EMI D+ RT +Y+ I+ N+ LF K
Sbjct 317 EAKAICEDGPQPQENTSEEDRQYFAGYAELSIHREMIGDTARTEAYRDFILQNRDLFADK 376
Query 169 V 169
V
Sbjct 377 V 377
> bbo:BBOV_III002510 17.m07242; hypothetical protein
Length=537
Score = 46.2 bits (108), Expect = 5e-05, Method: Composition-based stats.
Identities = 25/60 (41%), Positives = 33/60 (55%), Gaps = 6/60 (10%)
Query 110 KAAFSADWKQLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
K A S K +Q DD YF Y + +IH EM+ D VRT +Y+ I N+ LF+ KV
Sbjct 139 KTAKSQTKKCIQDDD------YFKGYKNLAIHREMVNDRVRTGAYESFIRLNKSLFKDKV 192
> ath:AT3G12270 PRMT3; PRMT3 (PROTEIN ARGININE METHYLTRANSFERASE
3); methyltransferase/ zinc ion binding; K11436 protein arginine
N-methyltransferase 3 [EC:2.1.1.-]
Length=601
Score = 45.1 bits (105), Expect = 1e-04, Method: Composition-based stats.
Identities = 19/39 (48%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 131 YFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
YF SY+ F IH EM+ D VRT +Y+ A++ N L G V
Sbjct 245 YFGSYSSFGIHREMLSDKVRTEAYRDALLKNPTLLNGSV 283
> xla:100137622 coactivator-associated arginine methyltransferase
1-b; K05931 histone-arginine methyltransferase CARM1 [EC:2.1.1.125]
Length=602
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 36/61 (59%), Gaps = 6/61 (9%)
Query 109 EKAAFSADWKQLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGK 168
EK+ FS ++ + SSA YF Y + S + M++D VRT +YQRAI+ N F+ K
Sbjct 104 EKSVFS------ERTEESSAVQYFQFYGYLSQQQNMMQDYVRTGTYQRAILQNHTDFKDK 157
Query 169 V 169
V
Sbjct 158 V 158
> tgo:TGME49_094270 arginine methyltransferase protein, putative
(EC:2.1.1.125); K05931 histone-arginine methyltransferase
CARM1 [EC:2.1.1.125]
Length=660
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 124 DISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
D S + YF Y + M++D+VRT +YQRAI++N+ F GK
Sbjct 206 DASVVETYFQYYGKMANQMNMLQDTVRTTTYQRAIVENRADFEGKT 251
> xla:494851 carm1; coactivator-associated arginine methyltransferase
1 (EC:2.1.1.125); K05931 histone-arginine methyltransferase
CARM1 [EC:2.1.1.125]
Length=602
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 36/61 (59%), Gaps = 6/61 (9%)
Query 109 EKAAFSADWKQLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGK 168
E++ FS ++ + SSA YF Y + S + M++D VRT +YQRAI+ N F+ K
Sbjct 104 ERSVFS------ERTEESSAVQYFQFYGYLSQQQNMMQDYVRTGTYQRAILQNHTDFKDK 157
Query 169 V 169
V
Sbjct 158 V 158
> mmu:99890 Prmt6, AW124876, BB233495, Hrmt1l6; protein arginine
N-methyltransferase 6 (EC:2.1.1.125); K11437 protein arginine
N-methyltransferase 6 [EC:2.1.1.-]
Length=378
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 131 YFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
Y+ Y+ S+HEEMI D VRT +Y+ I+ N RGK
Sbjct 50 YYECYSDVSVHEEMIADQVRTEAYRLGILKNWAALRGKT 88
> xla:447344 prmt6, MGC83989; protein arginine methyltransferase
6 (EC:2.1.1.125); K11437 protein arginine N-methyltransferase
6 [EC:2.1.1.-]
Length=340
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 19/37 (51%), Positives = 25/37 (67%), Gaps = 0/37 (0%)
Query 131 YFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRG 167
YF Y+ SIHEEMI D+VRT Y++AI+ N +G
Sbjct 18 YFQCYSDVSIHEEMIADTVRTNGYKQAILHNHCALQG 54
> dre:445251 carm1, PRMT4, si:dkey-204f11.63, zgc:100805; coactivator-associated
arginine methyltransferase 1 (EC:2.1.1.125);
K05931 histone-arginine methyltransferase CARM1 [EC:2.1.1.125]
Length=588
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/44 (47%), Positives = 28/44 (63%), Gaps = 0/44 (0%)
Query 126 SSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
SSA YF Y + S + M++D VRT +YQRAI+ N F+ KV
Sbjct 118 SSAVQYFQFYGYLSQQQNMMQDYVRTGTYQRAILQNHTDFKDKV 161
> hsa:10498 CARM1, PRMT4; coactivator-associated arginine methyltransferase
1 (EC:2.1.1.125); K05931 histone-arginine methyltransferase
CARM1 [EC:2.1.1.125]
Length=608
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 36/61 (59%), Gaps = 6/61 (9%)
Query 109 EKAAFSADWKQLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGK 168
E++ FS ++ + SSA YF Y + S + M++D VRT +YQRAI+ N F+ K
Sbjct 133 ERSVFS------ERTEESSAVQYFQFYGYLSQQQNMMQDYVRTGTYQRAILQNHTDFKDK 186
Query 169 V 169
+
Sbjct 187 I 187
> mmu:59035 Carm1, MGC46828, Prmt4; coactivator-associated arginine
methyltransferase 1 (EC:2.1.1.125); K05931 histone-arginine
methyltransferase CARM1 [EC:2.1.1.125]
Length=608
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 36/61 (59%), Gaps = 6/61 (9%)
Query 109 EKAAFSADWKQLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGK 168
E++ FS ++ + SSA YF Y + S + M++D VRT +YQRAI+ N F+ K
Sbjct 134 ERSVFS------ERTEESSAVQYFQFYGYLSQQQNMMQDYVRTGTYQRAILQNHTDFKDK 187
Query 169 V 169
+
Sbjct 188 I 188
> hsa:55170 PRMT6, FLJ10559, FLJ51477, HRMT1L6; protein arginine
methyltransferase 6 (EC:2.1.1.125); K11437 protein arginine
N-methyltransferase 6 [EC:2.1.1.-]
Length=375
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 131 YFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
Y+ Y+ S+HEEMI D VRT +Y+ I+ N RGK
Sbjct 47 YYECYSDVSVHEEMIADRVRTDAYRLGILRNWAALRGKT 85
> mmu:15468 Prmt2, AI504737, Hrmt1l1, MGC148245, MGC148246; protein
arginine N-methyltransferase 2 (EC:2.1.1.125); K11435
protein arginine N-methyltransferase 2 [EC:2.1.1.-]
Length=445
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query 118 KQLQQ---DDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
KQL++ +D + YF+SY +H EM+ D RT Y I+ N+ + KV
Sbjct 98 KQLEEYDPEDTWQDEEYFDSYGTLKLHLEMLADQPRTTKYHSVILQNKESLKDKV 152
> cpv:cgd8_3420 hypothetical protein
Length=324
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 125 ISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
+S+ D Y SY +H+ M++D R +Y+R+ +N+ LF+GK+
Sbjct 1 MSTLDTYSRSYDDLLVHQLMLQDVERVEAYKRSFEENKELFKGKI 45
> ath:AT3G06930 PRMT4B; protein arginine N-methyltransferase family
protein; K05931 histone-arginine methyltransferase CARM1
[EC:2.1.1.125]
Length=534
Score = 38.5 bits (88), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 126 SSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
SSA YF+ Y + M++D VRT +Y A+M+N F G+V
Sbjct 143 SSAKMYFHYYGQLLHQQNMLQDYVRTGTYYAAVMENHSDFAGRV 186
> ath:AT5G49020 PRMT4A; PRMT4A (PROTEIN ARGININE METHYLTRANSFERASE
4A); [myelin basic protein]-arginine N-methyltransferase/
histone-arginine N-methyltransferase/ methyltransferase/
protein heterodimerization/ protein homodimerization/ protein-arginine
omega-N asymmetric methyltransferase/ protein-arginine
omega-N monomethyltransferase; K05931 histone-arginine
methyltransferase CARM1 [EC:2.1.1.125]
Length=528
Score = 38.5 bits (88), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 27/44 (61%), Gaps = 0/44 (0%)
Query 126 SSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
+SA YF+ Y + M++D VRT +Y A+M+N+ F G+V
Sbjct 146 ASAKMYFHYYGQLLHQQNMLQDYVRTGTYHAAVMENRSDFSGRV 189
> pfa:PF08_0092 methyltransferase, putative
Length=912
Score = 37.7 bits (86), Expect = 0.017, Method: Composition-based stats.
Identities = 20/53 (37%), Positives = 27/53 (50%), Gaps = 1/53 (1%)
Query 118 KQLQQDDISSADY-YFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
K ++DI D YF+SY + IH MI D RT Y I N+ +F K+
Sbjct 492 KTYTKEDIHEIDKKYFDSYNYSDIHRTMILDKCRTQCYYDFINKNKEIFENKI 544
> dre:558841 prmt2, MGC158247, hrmt1l1, si:ch211-226c11.1, zgc:158247;
protein arginine methyltransferase 2 (EC:2.1.1.125);
K11435 protein arginine N-methyltransferase 2 [EC:2.1.1.-]
Length=408
Score = 37.7 bits (86), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 123 DDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
+D D YF +Y +H EM+ D RT +Y++ I+ N R KV
Sbjct 72 EDAWQDDEYFGNYGTLRLHLEMLSDKPRTETYRQVILSNSAALREKV 118
> cpv:cgd5_3770 arginine N-methyltransferase ; K11436 protein
arginine N-methyltransferase 3 [EC:2.1.1.-]
Length=665
Score = 37.7 bits (86), Expect = 0.019, Method: Composition-based stats.
Identities = 19/42 (45%), Positives = 25/42 (59%), Gaps = 3/42 (7%)
Query 131 YFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQ---HLFRGKV 169
YF+SY+ IH EMI D VRT +Y I D +LF+ K+
Sbjct 305 YFSSYSTLDIHREMILDKVRTDAYYNFITDPNNSINLFKDKI 346
> hsa:3275 PRMT2, HRMT1L1, MGC111373; protein arginine methyltransferase
2 (EC:2.1.1.125); K11435 protein arginine N-methyltransferase
2 [EC:2.1.1.-]
Length=433
Score = 36.6 bits (83), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 22/47 (46%), Gaps = 0/47 (0%)
Query 123 DDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
+D + YF SY +H EM+ D RT Y I+ N+ KV
Sbjct 94 EDTWQDEEYFGSYGTLKLHLEMLADQPRTTKYHSVILQNKESLTDKV 140
> xla:100499207 prmt2; protein arginine methyltransferase 2 (EC:2.1.1.125);
K11435 protein arginine N-methyltransferase 2
[EC:2.1.1.-]
Length=501
Score = 35.0 bits (79), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 23/46 (50%), Gaps = 0/46 (0%)
Query 123 DDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGK 168
+D + Y+ SY +H EM+ D RT +YQ I+ N GK
Sbjct 165 NDPWQDEEYYGSYKTLKLHLEMLSDVPRTMTYQNVILKNSSSLCGK 210
> ath:AT1G04870 PRMT10; PRMT10; [myelin basic protein]-arginine
N-methyltransferase/ histone-arginine N-methyltransferase/
methyltransferase/ protein-arginine omega-N asymmetric methyltransferase/
protein-arginine omega-N monomethyltransferase
Length=383
Score = 33.1 bits (74), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 2/44 (4%)
Query 128 ADY--YFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
DY YF +Y+ ++M+ D VR +Y A+ N+H F GK
Sbjct 30 VDYAQYFCTYSFLYHQKDMLSDRVRMDAYFNAVFQNKHHFEGKT 73
> cel:F54E2.3 ketn-1; KETtiN (Drosophila actin-binding) homolog
family member (ketn-1)
Length=4447
Score = 31.2 bits (69), Expect = 2.0, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 17/34 (50%), Gaps = 0/34 (0%)
Query 64 PSHAKGNGISANNGGSQTGNGSVRGGRGGRPAYA 97
P+ GI A NG ++T NGS G G +A
Sbjct 247 PTQVTNGGIKAANGSAKTANGSANGSANGSAVHA 280
> hsa:54809 SAMD9, C7orf5, DRIF1, FLJ20073, KIAA2004, NFTC, OEF1,
OEF2; sterile alpha motif domain containing 9
Length=1589
Score = 31.2 bits (69), Expect = 2.0, Method: Composition-based stats.
Identities = 17/63 (26%), Positives = 34/63 (53%), Gaps = 6/63 (9%)
Query 103 QKLSPEEKAAFSADWKQLQQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQ 162
Q+LSP+E+ AF K++++ + D+Y + ++E I++ VR I+ Q
Sbjct 855 QQLSPKEQRAFELKLKEIKEQHKNFEDFYSFMIMKTNFNKEYIENVVRN------ILKGQ 908
Query 163 HLF 165
++F
Sbjct 909 NIF 911
> tpv:TP01_0820 phosphatidylinositol 4-kinase; K00888 phosphatidylinositol
4-kinase [EC:2.7.1.67]
Length=1070
Score = 30.8 bits (68), Expect = 2.0, Method: Composition-based stats.
Identities = 18/81 (22%), Positives = 33/81 (40%), Gaps = 0/81 (0%)
Query 45 SVSILAQMESGQNHQKGALPSHAKGNGISANNGGSQTGNGSVRGGRGGRPAYAQSLQCQK 104
S ++ +++G H + +P N + N Q G+ S R P +S K
Sbjct 702 SPNLANSVQNGDTHVQNQVPDTNTPNQVGDTNTSVQNGDSSPNSNRDSTPVDKESSDSTK 761
Query 105 LSPEEKAAFSADWKQLQQDDI 125
SP + A + + K L ++
Sbjct 762 ESPRDGDAKTLNVKNLSPQEV 782
> dre:100003090 carm1l; coactivator-associated arginine methyltransferase
1, like; K05931 histone-arginine methyltransferase
CARM1 [EC:2.1.1.125]
Length=365
Score = 30.4 bits (67), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 29/49 (59%), Gaps = 8/49 (16%)
Query 121 QQDDISSADYYFNSYAHFSIHEEMIKDSVRTCSYQRAIMDNQHLFRGKV 169
Q+ + SSA YF + M++D +RT +YQ+A++ N+ F+ KV
Sbjct 119 QRSEDSSALQYFQ--------QNMLQDFLRTATYQKAMLLNEDDFKDKV 159
> dre:795013 phospholipase A2 receptor 1-like
Length=365
Score = 29.6 bits (65), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 33/116 (28%), Positives = 50/116 (43%), Gaps = 13/116 (11%)
Query 50 AQMESGQNHQKGALPSHAKGNGISANNGG-SQTG------NGSVRGGRGGRPAYAQSLQC 102
AQ+ +NH + + N + NNG S T N SV GGR AY +
Sbjct 155 AQLYCRENHTDLVSIRNEEENTLVMNNGNQSNTSFWIGLLNDSVDWRDGGRSAYRNWREK 214
Query 103 QKLSPEEKAAFSADWKQLQQDDISSADYY---FNSYAHFSIHEEMIKDSVRTCSYQ 155
S A SAD + + S+ ++Y + S+ H S+ E +D++ CS Q
Sbjct 215 TNHSMSYVAIQSADGRWFKA---SAGNFYPLCYKSFIHVSLAEMSWEDAMNYCSSQ 267
Lambda K H
0.318 0.130 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4222647260
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40