bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3151_orf1
Length=267
Score E
Sequences producing significant alignments: (Bits) Value
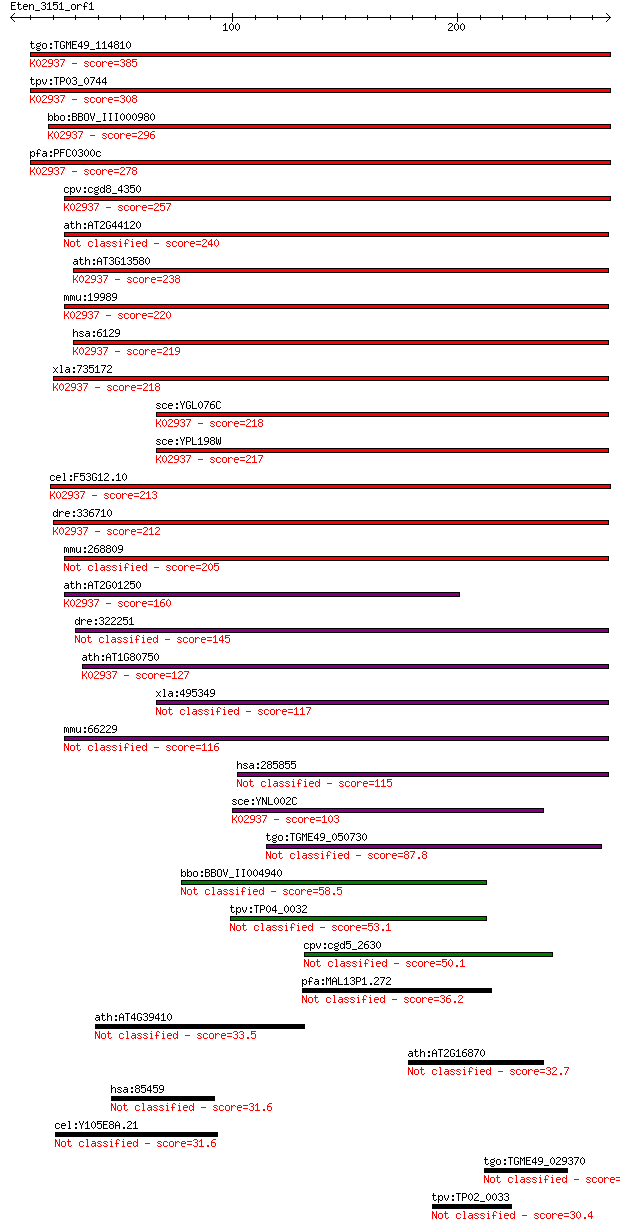
tgo:TGME49_114810 60S ribosomal protein L7, putative ; K02937 ... 385 9e-107
tpv:TP03_0744 60S ribosomal protein L7; K02937 large subunit r... 308 2e-83
bbo:BBOV_III000980 17.m07114; 60S ribosomal protein L7; K02937... 296 6e-80
pfa:PFC0300c 60S ribosomal protein L7, putative; K02937 large ... 278 1e-74
cpv:cgd8_4350 60S ribosomal protein L7 ; K02937 large subunit ... 257 3e-68
ath:AT2G44120 60S ribosomal protein L7 (RPL7C) 240 4e-63
ath:AT3G13580 60S ribosomal protein L7 (RPL7D); K02937 large s... 238 2e-62
mmu:19989 Rpl7, MGC102113, Rpl7a, Surf-3; ribosomal protein L7... 220 3e-57
hsa:6129 RPL7, MGC117326, humL7-1; ribosomal protein L7; K0293... 219 9e-57
xla:735172 rpl7, MGC130910; ribosomal protein L7; K02937 large... 218 2e-56
sce:YGL076C RPL7A; Rpl7ap; K02937 large subunit ribosomal prot... 218 3e-56
sce:YPL198W RPL7B; Rpl7bp; K02937 large subunit ribosomal prot... 217 4e-56
cel:F53G12.10 rpl-7; Ribosomal Protein, Large subunit family m... 213 5e-55
dre:336710 rpl7, bb02h10, mg:bb02h10, wu:fa93c11; ribosomal pr... 212 1e-54
mmu:268809 Gm5045, EG268809; predicted gene 5045 205 2e-52
ath:AT2G01250 60S ribosomal protein L7 (RPL7B); K02937 large s... 160 6e-39
dre:322251 rpl7l1, wu:fb54e11, zgc:66422; ribosomal protein L7... 145 2e-34
ath:AT1G80750 60S ribosomal protein L7 (RPL7A); K02937 large s... 127 5e-29
xla:495349 rpl7l1; ribosomal protein L7-like 1 117 3e-26
mmu:66229 Rpl7l1, 1500016H10Rik; ribosomal protein L7-like 1 116 7e-26
hsa:285855 RPL7L1, MGC62004, dJ475N16.4; ribosomal protein L7-... 115 2e-25
sce:YNL002C RLP7, RPL7; Nucleolar protein with similarity to l... 103 9e-22
tgo:TGME49_050730 ribosomal protein L7, putative 87.8 3e-17
bbo:BBOV_II004940 18.m06411; hypothetical protein 58.5 2e-08
tpv:TP04_0032 60S ribosomal protein L7 53.1 1e-06
cpv:cgd5_2630 60S ribosomal protein L7-B; Rp17bp; L30 RNA bind... 50.1 9e-06
pfa:MAL13P1.272 60S ribosomal protein L7-2, putative 36.2 0.12
ath:AT4G39410 WRKY13; WRKY13; transcription factor 33.5 0.75
ath:AT2G16870 disease resistance protein (TIR-NBS-LRR class), ... 32.7 1.4
hsa:85459 FLJ40913; KIAA1731 31.6 3.1
cel:Y105E8A.21 hypothetical protein 31.6 3.4
tgo:TGME49_029370 hypothetical protein 31.2 4.6
tpv:TP02_0033 hypothetical protein 30.4 6.3
> tgo:TGME49_114810 60S ribosomal protein L7, putative ; K02937
large subunit ribosomal protein L7e
Length=258
Score = 385 bits (989), Expect = 9e-107, Method: Compositional matrix adjust.
Identities = 179/258 (69%), Positives = 216/258 (83%), Gaps = 0/258 (0%)
Query 10 MADRFSARLLKPFEGVPLTEGAEKKLKRNAALAEARKKSLKAVREKRAANRHALKLRTYK 69
MADR+S +LL+P EGVP+ E + K+ R+ L K++ +A +K A RHALK+RTYK
Sbjct 1 MADRYSVKLLRPMEGVPVPERTKTKIARDVKLLNKMKQAKEAHAKKVHAQRHALKMRTYK 60
Query 70 YVKEYRQQREELIKNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQ 129
YV EYR++RE LIK KREA+A+GGFY+ E KV+ RIKGINKLAPKP+MIL+LFRLRQ
Sbjct 61 YVSEYRKERENLIKLKREAKAKGGFYKEPEAKVILATRIKGINKLAPKPKMILRLFRLRQ 120
Query 130 LHNGVFIRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNN 189
LHN VFI+VNKAT++ML+AVQPF+TYGYP+L TIR+LIYKRGYAKVG+PG+ RIRLQ N
Sbjct 121 LHNAVFIKVNKATIEMLKAVQPFITYGYPTLKTIRQLIYKRGYAKVGKPGAHSRIRLQAN 180
Query 190 DIIDKHLGKYGIHGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRP 249
DI+ +HLGKYGIHG+EDLVHEIYTCGP FKQAN FLW FKL+SPRKGF KRHG+ EPR
Sbjct 181 DIVSQHLGKYGIHGVEDLVHEIYTCGPYFKQANNFLWPFKLNSPRKGFTSKRHGYNEPRK 240
Query 250 GDWGNRQELINELVRRMC 267
GDWGNR+E+INELV+RM
Sbjct 241 GDWGNREEMINELVQRMI 258
> tpv:TP03_0744 60S ribosomal protein L7; K02937 large subunit
ribosomal protein L7e
Length=258
Score = 308 bits (788), Expect = 2e-83, Method: Compositional matrix adjust.
Identities = 144/258 (55%), Positives = 189/258 (73%), Gaps = 0/258 (0%)
Query 10 MADRFSARLLKPFEGVPLTEGAEKKLKRNAALAEARKKSLKAVREKRAANRHALKLRTYK 69
M RF R + + EG KK++ N L E + K K + E + K R K
Sbjct 1 MVSRFLVRSFRGLKKEECNEGLRKKMEHNEKLRELQAKKTKRLTEVKKKRVAGAKKRALK 60
Query 70 YVKEYRQQREELIKNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQ 129
Y KEY ++ EL++ +R+A+++G FY+ + KV F +RIKGINKL PKP++ L+LFRL Q
Sbjct 61 YAKEYVDRKRELVELRRKAKSEGSFYKEPDSKVAFVVRIKGINKLTPKPKLTLRLFRLLQ 120
Query 130 LHNGVFIRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNN 189
LHNGVF+++NKAT++ML+ V P+VTYGYPSLST+RKL+YKRGYAKVG+PG++QR+RL ++
Sbjct 121 LHNGVFVKLNKATVEMLKLVNPYVTYGYPSLSTVRKLLYKRGYAKVGKPGARQRVRLNSD 180
Query 190 DIIDKHLGKYGIHGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRP 249
D+I KHLGKYG+H +EDLVHE+YTCGP FKQ FLW FKL+ PRKGF KRHGF E R
Sbjct 181 DLISKHLGKYGVHSMEDLVHEVYTCGPHFKQVTNFLWPFKLNPPRKGFVAKRHGFSEARG 240
Query 250 GDWGNRQELINELVRRMC 267
GD GNR+ELIN+L+ RM
Sbjct 241 GDAGNREELINKLLARMV 258
> bbo:BBOV_III000980 17.m07114; 60S ribosomal protein L7; K02937
large subunit ribosomal protein L7e
Length=259
Score = 296 bits (757), Expect = 6e-80, Method: Compositional matrix adjust.
Identities = 141/253 (55%), Positives = 191/253 (75%), Gaps = 3/253 (1%)
Query 18 LLKPFEGVP---LTEGAEKKLKRNAALAEARKKSLKAVREKRAANRHALKLRTYKYVKEY 74
L+K F G+P E +K++RN L + +++ + E+R+ LK R Y KEY
Sbjct 7 LIKSFRGIPKEKCNETLLRKVERNEELRQLQQRKVAKHMERRSKRLAGLKKRALCYAKEY 66
Query 75 RQQREELIKNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGV 134
++ EL++ +R+A+A+GGFY+ A+ KVVF +RIKGINKL PKPR+IL+LFRL QLHNGV
Sbjct 67 LRRERELVELRRQAKAEGGFYKEADEKVVFVVRIKGINKLTPKPRLILKLFRLLQLHNGV 126
Query 135 FIRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDK 194
FI++NKAT++ML+ + P+VT+GYP+L+TIRKL+YKRGYAKVG G+ R+R+ ++ IID
Sbjct 127 FIKMNKATIEMLKLIAPYVTFGYPTLATIRKLLYKRGYAKVGRRGAWSRVRISDDQIIDD 186
Query 195 HLGKYGIHGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRPGDWGN 254
+LGK+GIHG+ED+VH+IYTCGP FK+ FLW FKL+ PRKGF KRHGF E R GD GN
Sbjct 187 NLGKHGIHGMEDIVHQIYTCGPKFKEVTNFLWPFKLNPPRKGFVAKRHGFTEARGGDAGN 246
Query 255 RQELINELVRRMC 267
R+ LINEL+ RM
Sbjct 247 REHLINELLLRMI 259
> pfa:PFC0300c 60S ribosomal protein L7, putative; K02937 large
subunit ribosomal protein L7e
Length=257
Score = 278 bits (712), Expect = 1e-74, Method: Compositional matrix adjust.
Identities = 139/258 (53%), Positives = 185/258 (71%), Gaps = 1/258 (0%)
Query 10 MADRFSARLLKPFEGVPLTEGAEKKLKRNAALAEARKKSLKAVREKRAANRHALKLRTYK 69
MADR+ ++ G +T KK+ +N L +A K +K ++ R+ L+ RT +
Sbjct 1 MADRYENQVENEL-GKSMTSLRAKKVNKNKELKQAAAKKIKTLKLINKKKRNDLRQRTLR 59
Query 70 YVKEYRQQREELIKNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQ 129
Y +EY +R+++I+ KREAR FYR AE KVVF IR+KG+NKL PK R + +L RL Q
Sbjct 60 YEEEYESERKKIIELKREARKNNCFYREAEKKVVFVIRLKGVNKLPPKVRSVFRLLRLLQ 119
Query 130 LHNGVFIRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNN 189
+HNGVF++VNKAT +ML+ V+P+VTYGYP+LST+RKL+YKRGY +VG+ R ++Q+N
Sbjct 120 VHNGVFVKVNKATKEMLKIVEPYVTYGYPTLSTVRKLLYKRGYVRVGKVRRYARKKIQDN 179
Query 190 DIIDKHLGKYGIHGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRP 249
I KHLGKY +HGIED+V+++YTCGP FK+ N FLW FKL PRKGFK KRH F EPRP
Sbjct 180 ADISKHLGKYNVHGIEDMVYQLYTCGPVFKKVNNFLWAFKLKPPRKGFKAKRHAFNEPRP 239
Query 250 GDWGNRQELINELVRRMC 267
GDWGNR+ INEL+ RM
Sbjct 240 GDWGNREAHINELINRMI 257
> cpv:cgd8_4350 60S ribosomal protein L7 ; K02937 large subunit
ribosomal protein L7e
Length=251
Score = 257 bits (657), Expect = 3e-68, Method: Compositional matrix adjust.
Identities = 128/243 (52%), Positives = 167/243 (68%), Gaps = 0/243 (0%)
Query 25 VPLTEGAEKKLKRNAALAEARKKSLKAVREKRAANRHALKLRTYKYVKEYRQQREELIKN 84
VP +E + KL+RN + + +K+ R K R K+ K K+Y++ + +I
Sbjct 9 VPPSESKKLKLERNMKIFSDFQSKIKSDRVKLNDLRKKAKIEALKLQKQYKEHVKSIIDL 68
Query 85 KREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGVFIRVNKATMQ 144
KR+A+ GG YR AEPKVVF IR+KGINKL+PK R I QLFRLRQ++NGVF+ +NKAT++
Sbjct 69 KRQAKQNGGLYRPAEPKVVFVIRLKGINKLSPKVRKIFQLFRLRQINNGVFLPLNKATIE 128
Query 145 MLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDKHLGKYGIHGI 204
ML+ + PFV YGYPS+S IRKL+YKRGY KVG+ GS QRIR+Q+N II K L G++GI
Sbjct 129 MLKVIDPFVAYGYPSISMIRKLLYKRGYLKVGKRGSYQRIRIQDNSIIQKKLASKGVYGI 188
Query 205 EDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRPGDWGNRQELINELVR 264
E +V E++ CG FK + LW FKLSSP GF K F E R GD GNR+ LIN+L+
Sbjct 189 EGMVRELFFCGEQFKAVTSLLWPFKLSSPNGGFVRKSTRFTEARGGDCGNRENLINKLID 248
Query 265 RMC 267
RMC
Sbjct 249 RMC 251
> ath:AT2G44120 60S ribosomal protein L7 (RPL7C)
Length=242
Score = 240 bits (612), Expect = 4e-63, Method: Compositional matrix adjust.
Identities = 124/242 (51%), Positives = 167/242 (69%), Gaps = 6/242 (2%)
Query 25 VPLTEGAEKKLKRNAALAEARKKSLKAVREKRAANRHALKLRTYKYVKEYRQQREELIKN 84
V + E KK+KR A A+K A ++K R + R +Y KEY ++ ELI+
Sbjct 6 VVVPESVLKKIKRQEEWALAKKDEAVAAKKKSVEARKLIFKRAEQYAKEYAEKDNELIRL 65
Query 85 KREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGVFIRVNKATMQ 144
KREA+ +GGFY E K++F IRI+GIN + PK + ILQL RLRQ+ NGVF++VNKAT+
Sbjct 66 KREAKLKGGFYVDPEAKLLFIIRIRGINAIDPKTKKILQLLRLRQIFNGVFLKVNKATVN 125
Query 145 MLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDKHLGKYGIHGI 204
ML+ V+P+VTYGYP+L ++++LIYKRGY K+ + QRI L +N I+D+ LGK+GI +
Sbjct 126 MLRRVEPYVTYGYPNLKSVKELIYKRGYGKL----NHQRIALTDNSIVDQALGKHGIICV 181
Query 205 EDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRPGDWGNRQELINELVR 264
EDL+HEI T GP FK+AN FLW F+L +P G K KR+ + E GD GNR+ INELVR
Sbjct 182 EDLIHEIMTVGPHFKEANNFLWPFQLKAPLGGLKKKRNHYVE--GGDAGNRENFINELVR 239
Query 265 RM 266
RM
Sbjct 240 RM 241
> ath:AT3G13580 60S ribosomal protein L7 (RPL7D); K02937 large
subunit ribosomal protein L7e
Length=244
Score = 238 bits (607), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 122/238 (51%), Positives = 168/238 (70%), Gaps = 6/238 (2%)
Query 29 EGAEKKLKRNAALAEARKKSLKAVREKRAANRHALKLRTYKYVKEYRQQREELIKNKREA 88
E KK KR A A+K+ L+A +++ A R + R +Y KEY+++ ELI+ KREA
Sbjct 12 ESVLKKRKREEEWALAKKQELEAAKKQNAEKRKLIFNRAKQYSKEYQEKERELIQLKREA 71
Query 89 RAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGVFIRVNKATMQMLQA 148
+ +GGFY E K++F IRI+GIN + PK + ILQL RLRQ+ NGVF++VNKAT+ ML+
Sbjct 72 KLKGGFYVDPEAKLLFIIRIRGINAIDPKTKKILQLLRLRQIFNGVFLKVNKATINMLRR 131
Query 149 VQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDKHLGKYGIHGIEDLV 208
V+P+VTYGYP+L ++++LIYKRG+ K+ + QR L +N I+D+ LGK+GI +EDL+
Sbjct 132 VEPYVTYGYPNLKSVKELIYKRGFGKL----NHQRTALTDNSIVDQGLGKHGIICVEDLI 187
Query 209 HEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRPGDWGNRQELINELVRRM 266
HEI T GP FK+AN FLW F+L +P G K KR+ + E GD GNR+ INELVRRM
Sbjct 188 HEIMTVGPHFKEANNFLWPFQLKAPLGGMKKKRNHYVE--GGDAGNRENFINELVRRM 243
> mmu:19989 Rpl7, MGC102113, Rpl7a, Surf-3; ribosomal protein
L7; K02937 large subunit ribosomal protein L7e
Length=270
Score = 220 bits (561), Expect = 3e-57, Method: Compositional matrix adjust.
Identities = 120/245 (48%), Positives = 161/245 (65%), Gaps = 11/245 (4%)
Query 25 VPLTEGAEKKLKRNAALAEARKKSLK---AVREKRAANRHALKLRTYKYVKEYRQQREEL 81
VP KK +RN AE + K L+ A++ R A R + + Y KEYRQ
Sbjct 33 VPAVPETLKKKRRN--FAELKVKRLRKKFALKTLRKARRKLIYEKAKHYHKEYRQMYRTE 90
Query 82 IKNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGVFIRVNKA 141
I+ R AR G FY AEPK+ F IRI+GIN ++PK R +LQL RLRQ+ NG F+++NKA
Sbjct 91 IRMARMARKAGNFYVPAEPKLAFVIRIRGINGVSPKVRKVLQLLRLRQIFNGTFVKLNKA 150
Query 142 TMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDKHLGKYGI 201
++ ML+ V+P++ +GYP+L ++ +LIYKRGY K+ +K+RI L +N +I + LGK+GI
Sbjct 151 SINMLRIVEPYIAWGYPNLKSVNELIYKRGYGKI----NKKRIALTDNSLIARSLGKFGI 206
Query 202 HGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRPGDWGNRQELINE 261
+EDL+HEIYT G FK+AN FLW FKLSSPR G K K F E GD GNR++ IN
Sbjct 207 ICMEDLIHEIYTVGKRFKEANNFLWPFKLSSPRGGMKKKTTHFVE--GGDAGNREDQINR 264
Query 262 LVRRM 266
L+RRM
Sbjct 265 LIRRM 269
> hsa:6129 RPL7, MGC117326, humL7-1; ribosomal protein L7; K02937
large subunit ribosomal protein L7e
Length=248
Score = 219 bits (557), Expect = 9e-57, Method: Compositional matrix adjust.
Identities = 122/252 (48%), Positives = 162/252 (64%), Gaps = 20/252 (7%)
Query 29 EGAEKKLKRNAALAEARKKS--------LKAVREK------RAANRHALKLRTYKYVKEY 74
EG E+K K A+ E KK +K +R+K R A R + + Y KEY
Sbjct 2 EGVEEKKKEVPAVPETLKKKRRNFAELKIKRLRKKFAQKMLRKARRKLIYEKAKHYHKEY 61
Query 75 RQQREELIKNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGV 134
RQ I+ R AR G FY AEPK+ F IRI+GIN ++PK R +LQL RLRQ+ NG
Sbjct 62 RQMYRTEIRMARMARKAGNFYVPAEPKLAFVIRIRGINGVSPKVRKVLQLLRLRQIFNGT 121
Query 135 FIRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDK 194
F+++NKA++ ML+ V+P++ +GYP+L ++ +LIYKRGY K+ +K+RI L +N +I +
Sbjct 122 FVKLNKASINMLRIVEPYIAWGYPNLKSVNELIYKRGYGKI----NKKRIALTDNALIAR 177
Query 195 HLGKYGIHGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRPGDWGN 254
LGKYGI +EDL+HEIYT G FK+AN FLW FKLSSPR G K K F E GD GN
Sbjct 178 SLGKYGIICMEDLIHEIYTVGKRFKEANNFLWPFKLSSPRGGMKKKTTHFVE--GGDAGN 235
Query 255 RQELINELVRRM 266
R++ IN L+RRM
Sbjct 236 REDQINRLIRRM 247
> xla:735172 rpl7, MGC130910; ribosomal protein L7; K02937 large
subunit ribosomal protein L7e
Length=246
Score = 218 bits (555), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 117/247 (47%), Positives = 159/247 (64%), Gaps = 8/247 (3%)
Query 20 KPFEGVPLTEGAEKKLKRNAALAEARKKSLKAVREKRAANRHALKLRTYKYVKEYRQQRE 79
K VP E K+ K+ AA R K + A ++ R R + R Y KEYRQ
Sbjct 7 KKLPSVP--ESLLKRRKQFAAAKLKRVKKILAAKKLRKTKRKVIFKRAESYYKEYRQIYR 64
Query 80 ELIKNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGVFIRVN 139
+ ++ R AR G FY AEPK+ F IRI+GIN ++PK R +LQL RLRQ+ NG F+++N
Sbjct 65 KEVRLARIARKAGNFYVPAEPKLAFVIRIRGINGVSPKVRKVLQLLRLRQIFNGTFVKLN 124
Query 140 KATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDKHLGKY 199
KA++ ML+ V+P++ +GYP+L ++R+LIYKRGY K+ QRI + +N +I+K+LGK
Sbjct 125 KASINMLRLVEPYIAWGYPNLKSVRQLIYKRGYLKI----KNQRIPMTDNSLIEKYLGKK 180
Query 200 GIHGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRPGDWGNRQELI 259
G+ +EDL+HEIYT G FK AN FLW FKLSSPR G K F E GD GNR++ I
Sbjct 181 GLMCVEDLIHEIYTVGKNFKAANNFLWPFKLSSPRGGMNKKTTHFVE--GGDAGNREDQI 238
Query 260 NELVRRM 266
N ++RRM
Sbjct 239 NRMIRRM 245
> sce:YGL076C RPL7A; Rpl7ap; K02937 large subunit ribosomal protein
L7e
Length=244
Score = 218 bits (554), Expect = 3e-56, Method: Compositional matrix adjust.
Identities = 103/201 (51%), Positives = 144/201 (71%), Gaps = 4/201 (1%)
Query 66 RTYKYVKEYRQQREELIKNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLF 125
R Y KEY +I+ KR+A+A G +Y A+ K+VF +RIKGINK+ PKPR +LQL
Sbjct 47 RNAAYQKEYETAERNIIQAKRDAKAAGSYYVEAQHKLVFVVRIKGINKIPPKPRKVLQLL 106
Query 126 RLRQLHNGVFIRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIR 185
RL ++++G F++V KAT+++L+ ++P+V YGYPS STIR+L+YKRG+ K+ +KQR+
Sbjct 107 RLTRINSGTFVKVTKATLELLKLIEPYVAYGYPSYSTIRQLVYKRGFGKI----NKQRVP 162
Query 186 LQNNDIIDKHLGKYGIHGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFC 245
L +N II+ +LGKYGI I+DL+HEI T GP FKQAN FLW FKLS+P G+ R
Sbjct 163 LSDNAIIEANLGKYGILSIDDLIHEIITVGPHFKQANNFLWPFKLSNPSGGWGVPRKFKH 222
Query 246 EPRPGDWGNRQELINELVRRM 266
+ G +GNR+E IN+LV+ M
Sbjct 223 FIQGGSFGNREEFINKLVKSM 243
> sce:YPL198W RPL7B; Rpl7bp; K02937 large subunit ribosomal protein
L7e
Length=244
Score = 217 bits (552), Expect = 4e-56, Method: Compositional matrix adjust.
Identities = 103/201 (51%), Positives = 144/201 (71%), Gaps = 4/201 (1%)
Query 66 RTYKYVKEYRQQREELIKNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLF 125
R Y KEY +I+ KR+A+A G +Y A+ K+VF +RIKGINK+ PKPR +LQL
Sbjct 47 RNAAYQKEYETAERNIIQAKRDAKAAGSYYVEAQHKLVFVVRIKGINKIPPKPRKVLQLL 106
Query 126 RLRQLHNGVFIRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIR 185
RL ++++G F++V KAT+++L+ ++P+V YGYPS STIR+L+YKRG+ K+ +KQR+
Sbjct 107 RLTRINSGTFVKVTKATLELLKLIEPYVAYGYPSYSTIRQLVYKRGFGKI----NKQRVP 162
Query 186 LQNNDIIDKHLGKYGIHGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFC 245
L +N II+ +LGKYGI I+DL+HEI T GP FKQAN FLW FKLS+P G+ R
Sbjct 163 LSDNAIIEANLGKYGILSIDDLIHEIITVGPHFKQANNFLWPFKLSNPSGGWGVPRKFKH 222
Query 246 EPRPGDWGNRQELINELVRRM 266
+ G +GNR+E IN+LV+ M
Sbjct 223 FIQGGSFGNREEFINKLVKAM 243
> cel:F53G12.10 rpl-7; Ribosomal Protein, Large subunit family
member (rpl-7); K02937 large subunit ribosomal protein L7e
Length=244
Score = 213 bits (542), Expect = 5e-55, Method: Compositional matrix adjust.
Identities = 115/252 (45%), Positives = 164/252 (65%), Gaps = 11/252 (4%)
Query 19 LKPFEGVPLTEGAEKKLKRNAALAEARKKSLK---AVREKRAANRHALKLRTYKYVKEYR 75
+ P + VP + E LKR A+AR K+ + V K + R KYV+EYR
Sbjct 1 MAPTKKVP--QVPETVLKRRKQRADARTKAAQHKVTVAAKNKEKKTQYFKRAEKYVQEYR 58
Query 76 QQREELIKNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGVF 135
++E ++ KREA A+G FY AE KV F +RI+GIN+L PKPR LQ+ RLRQ++NGVF
Sbjct 59 NAQKEGLRLKREAEAKGDFYVPAEHKVAFVVRIRGINQLHPKPRKALQILRLRQINNGVF 118
Query 136 IRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDKH 195
+++NKAT+ +L+ ++P+V +GYP+ TI L+YKRGYAKV R+ + +N I+++
Sbjct 119 VKLNKATLPLLRIIEPYVAWGYPNNKTIHDLLYKRGYAKV----DGNRVPITDNTIVEQS 174
Query 196 LGKYGIHGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRPGDWGNR 255
LGK+ I +EDL HEI T GP FK+A FLW FKL++P G+ K + F E GD+GNR
Sbjct 175 LGKFNIICLEDLAHEIATVGPHFKEATNFLWPFKLNNPTGGWTKKTNHFVE--GGDFGNR 232
Query 256 QELINELVRRMC 267
++ IN L+R+M
Sbjct 233 EDQINNLLRKMV 244
> dre:336710 rpl7, bb02h10, mg:bb02h10, wu:fa93c11; ribosomal
protein L7; K02937 large subunit ribosomal protein L7e
Length=246
Score = 212 bits (540), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 120/247 (48%), Positives = 158/247 (63%), Gaps = 8/247 (3%)
Query 20 KPFEGVPLTEGAEKKLKRNAALAEARKKSLKAVREKRAANRHALKLRTYKYVKEYRQQRE 79
K VP E K+ +R AA+ R K KA ++ R + R Y KEY+Q
Sbjct 7 KKIPSVP--ESLLKRRQRFAAIRAVRLKKAKADKKASKVTRKLIFKRAEAYHKEYKQLYR 64
Query 80 ELIKNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGVFIRVN 139
I+ R AR G +Y AEPK+ F IRI+GIN ++PK R +LQL RLRQ+ NGVF+++N
Sbjct 65 REIRMSRMARKAGNYYVPAEPKLAFVIRIRGINGVSPKVRKVLQLLRLRQIFNGVFVKLN 124
Query 140 KATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDKHLGKY 199
KA++ ML+ +P++ +GYP+L ++R+LIYKRG+ K+ KQRI L +N +I+K LG+
Sbjct 125 KASINMLRIAEPYIAWGYPNLKSVRELIYKRGFGKI----KKQRIALTDNSLIEKTLGQC 180
Query 200 GIHGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRPGDWGNRQELI 259
GI IEDL+HEIYT G FK AN FLW FKLSSPR G K F E GD GNR++ I
Sbjct 181 GIICIEDLIHEIYTVGKNFKAANNFLWPFKLSSPRGGMNKKTTHFVE--GGDAGNREDQI 238
Query 260 NELVRRM 266
N LVRRM
Sbjct 239 NRLVRRM 245
> mmu:268809 Gm5045, EG268809; predicted gene 5045
Length=270
Score = 205 bits (521), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 115/245 (46%), Positives = 157/245 (64%), Gaps = 11/245 (4%)
Query 25 VPLTEGAEKKLKRNAALAEARKKSLK---AVREKRAANRHALKLRTYKYVKEYRQQREEL 81
VP KK +RN AE + K L A++ R A R + + Y KEYRQ
Sbjct 33 VPAVPETLKKKRRN--FAELKVKCLPKKFALKTLRKARRKLIYEKAKCYHKEYRQMYCTE 90
Query 82 IKNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGVFIRVNKA 141
I+ R AR G FY +AEPK+ F IRI+ IN ++PK R + QL RLRQ+ NG F ++NKA
Sbjct 91 IRMARMARKAGSFYVLAEPKMAFVIRIRDINGVSPKVRKVPQLLRLRQIFNGTFAKLNKA 150
Query 142 TMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDKHLGKYGI 201
++ ML+ V+P++ +GYP+L ++ +LIYKRGY K+ +K++I L +N +I + LGK+GI
Sbjct 151 SINMLRIVEPYIAWGYPNLKSVNELIYKRGYGKI----NKKQIALTDNSLIARSLGKFGI 206
Query 202 HGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRPGDWGNRQELINE 261
+EDL+HEI T G FK+AN FLW FKLSSP+ G K K F E GD GNR++ IN
Sbjct 207 ICMEDLIHEISTVGKRFKEANNFLWPFKLSSPQGGMKKKTTHFVE--GGDAGNREDQINR 264
Query 262 LVRRM 266
L+RRM
Sbjct 265 LIRRM 269
> ath:AT2G01250 60S ribosomal protein L7 (RPL7B); K02937 large
subunit ribosomal protein L7e
Length=184
Score = 160 bits (404), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 83/176 (47%), Positives = 121/176 (68%), Gaps = 4/176 (2%)
Query 25 VPLTEGAEKKLKRNAALAEARKKSLKAVREKRAANRHALKLRTYKYVKEYRQQREELIKN 84
V + E KK KR A +K++++A ++K A NR + R +Y KEY ++ +ELI
Sbjct 6 VVVPESVLKKRKREEEWALEKKQNVEAAKKKNAENRKLIFKRAEQYSKEYAEKEKELISL 65
Query 85 KREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGVFIRVNKATMQ 144
KREA+ +GGFY E K++F IRI+GIN + PK + ILQL RLRQ+ NGVF++VNKATM
Sbjct 66 KREAKLKGGFYVDPEAKLLFIIRIRGINAIDPKTKKILQLLRLRQIFNGVFLKVNKATMN 125
Query 145 MLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDKHLGKYG 200
ML+ V+P+VTYG+P+L ++++LIYKRGY K+ QRI L +N I+++ +G
Sbjct 126 MLRRVEPYVTYGFPNLKSVKELIYKRGYGKLNH----QRIALTDNSIVEQPTTSFG 177
> dre:322251 rpl7l1, wu:fb54e11, zgc:66422; ribosomal protein
L7-like 1
Length=247
Score = 145 bits (365), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 88/252 (34%), Positives = 150/252 (59%), Gaps = 25/252 (9%)
Query 30 GAEKKLKRNAALAEARKKSLKAVREKRAANRHALKLRTYKYVKE-----YRQQREELIKN 84
G EK +K ++K+ +A++ AN+ L L+ K V++ +++ E L +
Sbjct 5 GTEKVIKLVPENLLKKRKAYQAIK----ANQAKLALQQKKRVQKGIPFKFKRLEEFLQNS 60
Query 85 KREARAQGGFYRV----------AEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGV 134
+R+ R + R+ A+ + F +RI+ I ++PK ++Q+ RLR++ +G
Sbjct 61 RRKHRDETRLARLKKRPPPPMPPAKNSLAFAVRIREIKGVSPKVMNVIQMLRLRKIFSGT 120
Query 135 FIRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDK 194
F++V+K ++ ML+ V+P+V +GYP+L ++R+LI KRG KV K++I L +N +I++
Sbjct 121 FVKVSKHSINMLKTVEPYVAWGYPNLKSVRELILKRGQGKV----DKRKIPLTDNTLIEQ 176
Query 195 HLGKYGIHGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRPGDWGN 254
HLGKYGI +EDL+HEIY+ G F+ N FLW F+LS PR + K G + G+ G
Sbjct 177 HLGKYGIICLEDLIHEIYSAGKNFRVVNNFLWPFRLSVPRHAARDK-VGLMKD-VGNPGP 234
Query 255 RQELINELVRRM 266
R E IN ++R++
Sbjct 235 RAEDINTVIRKL 246
> ath:AT1G80750 60S ribosomal protein L7 (RPL7A); K02937 large
subunit ribosomal protein L7e
Length=247
Score = 127 bits (318), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 82/236 (34%), Positives = 128/236 (54%), Gaps = 9/236 (3%)
Query 33 KKLKRNAALAEARKKSLKAVREKRAANRHALKLRTYKYVKEYRQQREELIKNKREARAQG 92
KK K LA RKK L+ + + + R +V E+R + ++I+ K+ +
Sbjct 18 KKRKNRDELAFIRKKQLELGNSGKKKKKVSDIKRPEDFVHEFRAKEVDMIRMKQRVKRPK 77
Query 93 GFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGVFIRVNKATMQMLQAVQPF 152
+ +VF IRI+G N + PK + IL +L+ + GVF + + Q L VQP+
Sbjct 78 SSPPPVKSDLVFIIRIQGKNDMHPKTKRILNNLQLKSVFTGVFAKATDSLFQKLLKVQPY 137
Query 153 VTYGYPSLSTIRKLIYKRGYAKV-GEPGSKQRIRLQNNDIIDKHLGKYGIHGIEDLVHEI 211
VTYGYP+ +++ LIYK+G + G P + L +N+II++ LG++ I GIEDLV+EI
Sbjct 138 VTYGYPNDKSVKDLIYKKGCTIIEGNP-----VPLTDNNIIEQALGEHKILGIEDLVNEI 192
Query 212 YTCGPAFKQANAFLWTFKLSSPRKG-FKCKRHGFCEPRPGDWGNRQELINELVRRM 266
G F++ FL KL+ P K+ F E GD GNR++ IN+L+ +M
Sbjct 193 ARVGDHFREVMRFLGPLKLNKPVADVLHRKKQVFSE--GGDTGNREDKINDLISKM 246
> xla:495349 rpl7l1; ribosomal protein L7-like 1
Length=246
Score = 117 bits (294), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 63/201 (31%), Positives = 119/201 (59%), Gaps = 6/201 (2%)
Query 66 RTYKYVKEYRQQREELIKNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLF 125
R +V++ ++ + ++ KR Q + + + F +RI I+ ++ + L+
Sbjct 51 RLESFVRDSHRKLRDDVRLKRMRIYQKKIPGIDKHGLAFVVRITKIDGVSKAVQDALKKL 110
Query 126 RLRQLHNGVFIRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIR 185
RL ++ G F++V +T +++ V+P+V +G+P+L +IR+L+ KRG+ + + +R+
Sbjct 111 RLGKIFAGAFVKVTPSTENLIRVVEPYVAWGFPNLKSIRELVLKRGHFR----KNGRRVA 166
Query 186 LQNNDIIDKHLGKYGIHGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFC 245
L +N++I++ LGK+GI +ED++HE+YT G F + N+FL F+LS R R G+
Sbjct 167 LTDNNVIEEQLGKFGIICLEDVIHELYTTGEHFSEVNSFLCPFRLSVSRHA-AVNRKGYL 225
Query 246 EPRPGDWGNRQELINELVRRM 266
GD GNR IN+L+R++
Sbjct 226 S-EVGDPGNRGAGINQLIRQL 245
> mmu:66229 Rpl7l1, 1500016H10Rik; ribosomal protein L7-like 1
Length=246
Score = 116 bits (291), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 79/244 (32%), Positives = 142/244 (58%), Gaps = 10/244 (4%)
Query 25 VPLT-EGAEKKLKRNAAL-AEARKKSLKAVREKRAANRHALKLRTYKYVKEYRQQREELI 82
+PL E KK K AL A K++L A RE++ +L ++ + +RQQR+++
Sbjct 10 IPLVPENLLKKRKAYQALKATQAKQALLAKRERKGKQFRFRRLESFVH-DSWRQQRDKVR 68
Query 83 KNKREARAQGGFYRVAEPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGVFIRVNKAT 142
+ E + + P + F IR++ I ++ + + L++L +GVF++V +
Sbjct 69 VQRLEVKPRALEVPDKHP-LAFVIRMERIEGVSLLVKSTIMKLGLKKLFSGVFVKVTPQS 127
Query 143 MQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDKHLGKYGIH 202
++ML+ V+P+VT+G+P+L ++R+LI KRG AK+ + + + L +N +I++HLG++G+
Sbjct 128 VRMLRTVEPYVTWGFPNLKSVRELILKRGQAKI----NNKTVPLTDNTVIEEHLGRFGVI 183
Query 203 GIEDLVHEIYTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPRPGDWGNRQELINEL 262
+EDL+HEI G F++ ++FL F LS R + R GF G G R + IN+L
Sbjct 184 CLEDLIHEIAFPGKHFQEVSSFLCPFLLSVARHATR-NRVGF-RKEMGSPGYRGDRINQL 241
Query 263 VRRM 266
+R++
Sbjct 242 IRQL 245
> hsa:285855 RPL7L1, MGC62004, dJ475N16.4; ribosomal protein L7-like
1
Length=246
Score = 115 bits (287), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 63/165 (38%), Positives = 105/165 (63%), Gaps = 6/165 (3%)
Query 102 VVFCIRIKGINKLAPKPRMILQLFRLRQLHNGVFIRVNKATMQMLQAVQPFVTYGYPSLS 161
+ F +RI+ I+ ++ + + RL+++ +GVF++V ++ML+ V+P+VT+G+P+L
Sbjct 87 LAFVVRIERIDGVSLLVQRTIARLRLKKIFSGVFVKVTPQNLKMLRIVEPYVTWGFPNLK 146
Query 162 TIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDKHLGKYGIHGIEDLVHEIYTCGPAFKQA 221
++R+LI KRG AKV + I L +N +I++HLGK+G+ +EDL+HEI G F++
Sbjct 147 SVRELILKRGQAKV----KNKTIPLTDNTVIEEHLGKFGVICLEDLIHEIAFPGKHFQEI 202
Query 222 NAFLWTFKLSSPRKGFKCKRHGFCEPRPGDWGNRQELINELVRRM 266
+ FL F LS R K R GF + G G R E IN+L+R++
Sbjct 203 SWFLCPFHLSVARHATK-NRVGFLKEM-GTPGYRGERINQLIRQL 245
> sce:YNL002C RLP7, RPL7; Nucleolar protein with similarity to
large ribosomal subunit L7 proteins; constituent of 66S pre-ribosomal
particles; plays an essential role in processing
of precursors to the large ribosomal subunit RNAs; K02937 large
subunit ribosomal protein L7e
Length=322
Score = 103 bits (256), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 55/143 (38%), Positives = 83/143 (58%), Gaps = 6/143 (4%)
Query 100 PKVVFCIRIKG---INKLAPKPRMILQLFRLRQLHNGVFIRVNKATMQMLQAVQPFVTYG 156
P ++F +R++G +N + K IL L RL + + GVF+++ K +L+ + P+V G
Sbjct 139 PALLFIVRVRGPLAVN-IPNKAFKILSLLRLVETNTGVFVKLTKNVYPLLKVIAPYVVIG 197
Query 157 YPSLSTIRKLIYKRG--YAKVGEPGSKQRIRLQNNDIIDKHLGKYGIHGIEDLVHEIYTC 214
PSLS+IR LI KRG K I L +N+I+++ LG +GI +ED++HEI T
Sbjct 198 KPSLSSIRSLIQKRGRIIYKGENEAEPHEIVLNDNNIVEEQLGDHGIICVEDIIHEIATM 257
Query 215 GPAFKQANAFLWTFKLSSPRKGF 237
G +F N FL FKL+ GF
Sbjct 258 GESFSVCNFFLQPFKLNREVSGF 280
> tgo:TGME49_050730 ribosomal protein L7, putative
Length=267
Score = 87.8 bits (216), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 50/151 (33%), Positives = 85/151 (56%), Gaps = 13/151 (8%)
Query 115 APKPRMILQLFRLRQLHNGVFIRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAK 174
+P+ + L+ LR+ G +R ++AT Q ++ + PFV YGYP++ T+RKL RG +
Sbjct 124 SPEVQAGLKRLGLRKPFWGTVLRNDEATSQQMRLLDPFVFYGYPTIQTLRKLFLTRGCLR 183
Query 175 VGEPGSKQRIRLQNNDIIDKHLGKYGIHGIEDLVHEIYTCGPAFKQANAFLWTFKLSSPR 234
+ + K+ L +N +++HLG YG+ +ED+V E++T G F L F+LS+ +
Sbjct 184 IND---KETTSLTDNAKVEEHLGAYGMLCVEDVVQELWTAGRHFDDIKQHLCAFQLSNLK 240
Query 235 K--GFKCKRHGFCEPRPGDWGNRQELINELV 263
K G +R+ F GN +E IN+ +
Sbjct 241 KVEGLYARRNEF--------GNMREAINKKI 263
> bbo:BBOV_II004940 18.m06411; hypothetical protein
Length=223
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 40/137 (29%), Positives = 70/137 (51%), Gaps = 6/137 (4%)
Query 77 QREELIKNKREARAQGGFYRVAEPKVVFCIRIK-GINKLAPKPRMILQLFRLRQLHNGVF 135
QR+ IK + + Y ++F IR K + A R L++ L +G F
Sbjct 60 QRDRDIKRTKALEKKPKVYAKQPYNLLFVIRNKRSVESTAL--RSTLRVMGLWHYGSGRF 117
Query 136 IRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDKH 195
++++++ L V+PFV YG P+L +R L++KRG ++ + G+ I N +++ H
Sbjct 118 FSNSESSLRDLDLVKPFVFYGTPTLKNVRNLLHKRGTVRL-DNGADTLI--SGNSVVEDH 174
Query 196 LGKYGIHGIEDLVHEIY 212
LG G+ + D+V IY
Sbjct 175 LGDTGLLCVADVVDAIY 191
> tpv:TP04_0032 60S ribosomal protein L7
Length=236
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/114 (28%), Positives = 63/114 (55%), Gaps = 5/114 (4%)
Query 99 EPKVVFCIRIKGINKLAPKPRMILQLFRLRQLHNGVFIRVNKATMQMLQAVQPFVTYGYP 158
E K++F +R N +P + IL+ L + ++G F+ + ++++ V PFV YG P
Sbjct 82 EYKLLFVVR-NSRNVESPICKDILKTLGLTRTNSGRFMSNSVPNLELINKVLPFVYYGTP 140
Query 159 SLSTIRKLIYKRGYAKVGEPGSKQRIRLQNNDIIDKHLGKYGIHGIEDLVHEIY 212
+L ++ L++KRG + + + I N +I+++ YGI+ I +L+ IY
Sbjct 141 TLKHVKDLLHKRG--TIYQHNKAEMI--SGNLVIEENFSIYGIYSIAELIETIY 190
> cpv:cgd5_2630 60S ribosomal protein L7-B; Rp17bp; L30 RNA binding
domain
Length=268
Score = 50.1 bits (118), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 32/130 (24%), Positives = 68/130 (52%), Gaps = 20/130 (15%)
Query 132 NGVFIRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRG---YAKVGEPGSKQ------ 182
+ V + T+++L+ V+P+V YG+P+L+ ++ L++K+G + V G +
Sbjct 121 SAVILSHTDETIRVLREVKPYVFYGFPNLNLLKTLVFKKGAFTHQNVQSKGQSRAKLSEK 180
Query 183 ---RIRLQNNDIIDKHLGKYGIHGIEDLVHEIYT-CGP-----AFKQANAFLWTFKLSSP 233
++ L +N+I++ LG+ G+ ED+V ++ C F+ + L F+L +
Sbjct 181 KQGKVLLTDNNIVEDRLGEIGVLCTEDIVTGLWNGCETQQDKEIFEAITSNLAPFQLCNL 240
Query 234 RK--GFKCKR 241
++ GF K+
Sbjct 241 KRAEGFDAKK 250
> pfa:MAL13P1.272 60S ribosomal protein L7-2, putative
Length=252
Score = 36.2 bits (82), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 20/84 (23%), Positives = 38/84 (45%), Gaps = 4/84 (4%)
Query 131 HNGVFIRVNKATMQMLQAVQPFVTYGYPSLSTIRKLIYKRGYAKVGEPGSKQRIRLQNND 190
++G+ + + M+ L ++P++ YGY L+ KR Y + Q R +N
Sbjct 122 YDGIILINTEENMKKLYVIKPYICYGYIKKYNFYNLMEKRLYIR----DQDQIKRCDSNK 177
Query 191 IIDKHLGKYGIHGIEDLVHEIYTC 214
+I++ K GI+ I+ C
Sbjct 178 MIEQIFSKEGIYSFRSFCDYIFEC 201
> ath:AT4G39410 WRKY13; WRKY13; transcription factor
Length=304
Score = 33.5 bits (75), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 46/93 (49%), Gaps = 6/93 (6%)
Query 39 AALAEARKKSLKAVREKRAANRHALKLRTYKYVKEYRQQREELIKNKREARAQGGFYRVA 98
++L + K+ + VRE R + ++ +R+ ++++KN + R+ +YR
Sbjct 193 SSLKMKKLKTRRKVREPRFCFKTLSEVDVLDDGYRWRKYGQKVVKNTQHPRS---YYRCT 249
Query 99 EPKVVFCIRIKGINKLAPKPRMILQLFRLRQLH 131
+ K C K + +LA PRM++ + R LH
Sbjct 250 QDK---CRVKKRVERLADDPRMVITTYEGRHLH 279
> ath:AT2G16870 disease resistance protein (TIR-NBS-LRR class),
putative
Length=1109
Score = 32.7 bits (73), Expect = 1.4, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 34/62 (54%), Gaps = 12/62 (19%)
Query 178 PGSKQRIRLQNNDIIDKHLGKYGIHGIEDLVHEIYTCGPAFKQANAFL--WTFKLSSPRK 235
PGS+ + +N +I+ +H GI+++ H + P+ ++A L + FK SSPR
Sbjct 317 PGSRVIVTTENKEILQRH-------GIDNMYHVGF---PSDEKAMEILCGYAFKQSSPRP 366
Query 236 GF 237
GF
Sbjct 367 GF 368
> hsa:85459 FLJ40913; KIAA1731
Length=2601
Score = 31.6 bits (70), Expect = 3.1, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Query 46 KKSLKAVREKRAANRHALKLRTYKYVKEYRQQREELIKNKREARAQ 91
+ S R+ A RH LR Y + E +QQ+EE K K+EA AQ
Sbjct 2536 RASFPEDRKTTQALRHQRGLRLYNQLAEVKQQKEE--KTKQEAYAQ 2579
> cel:Y105E8A.21 hypothetical protein
Length=934
Score = 31.6 bits (70), Expect = 3.4, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 35/72 (48%), Gaps = 5/72 (6%)
Query 21 PFEGVPLTEGAEKKLKRNAALAEARKKSLKAVREKRAANRHALKLRTYKYVKEYRQQREE 80
P G L+ G + +L +A A + L +R ANR+ + L+ + +KE + E
Sbjct 772 PAVGTLLSSGIDGRL-----VASATGRFLGNMRHSYFANRNVMALKRRRILKESQTMEEY 826
Query 81 LIKNKREARAQG 92
L + K E A+G
Sbjct 827 LKEVKMEGSAEG 838
> tgo:TGME49_029370 hypothetical protein
Length=1626
Score = 31.2 bits (69), Expect = 4.6, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Query 212 YTCGPAFKQANAFLWTFKLSSPRKGFKCKRHGFCEPR 248
Y G F +A AF WT K+ S + F K+HGF + R
Sbjct 1118 YVHGVRF-EAEAFAWTAKIGSGSRRFLVKKHGFHKSR 1153
> tpv:TP02_0033 hypothetical protein
Length=775
Score = 30.4 bits (67), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 10/35 (28%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 189 NDIIDKHLGKYGIHGIEDLVHEIYTCGPAFKQANA 223
N + DK L + + + DL+ ++ + PAF+++N+
Sbjct 462 NYVWDKELSSWRFNSVPDLIRDLLSVNPAFRESNS 496
Lambda K H
0.323 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9954723792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40