bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3208_orf2
Length=212
Score E
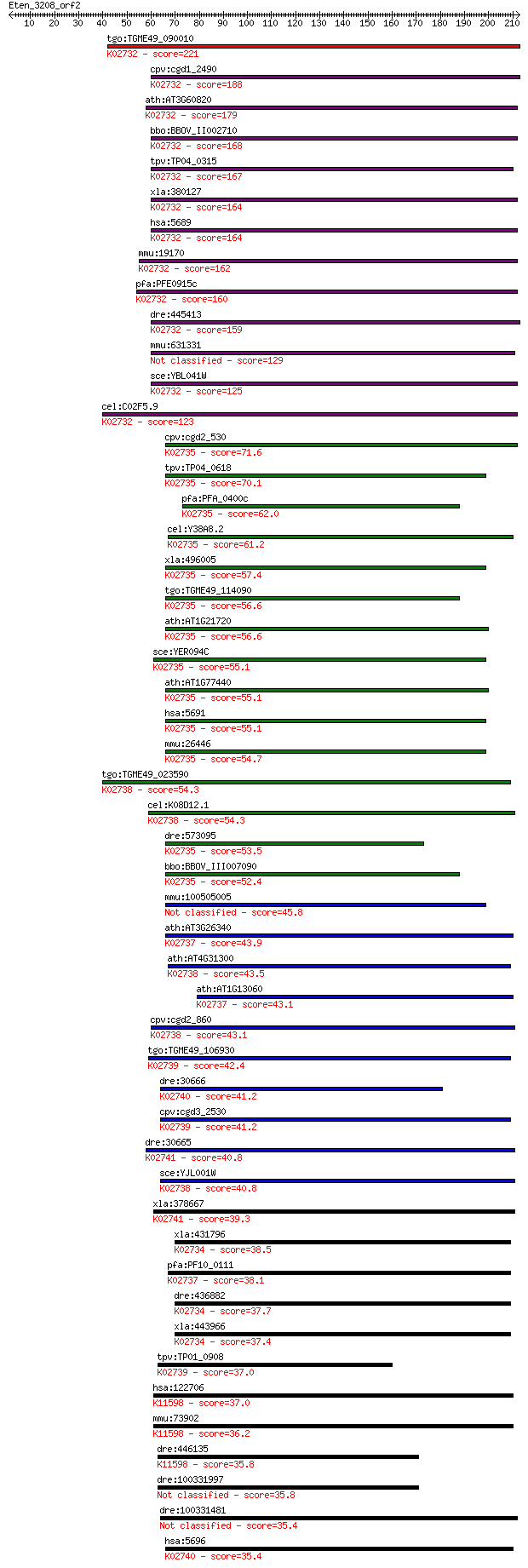
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_090010 proteasome subunit beta type 1, putative (EC... 221 2e-57
cpv:cgd1_2490 proteasome subunit beta type 1 ; K02732 20S prot... 188 1e-47
ath:AT3G60820 PBF1; PBF1; peptidase/ threonine-type endopeptid... 179 5e-45
bbo:BBOV_II002710 18.m06221; proteasome A-type and B-type fami... 168 1e-41
tpv:TP04_0315 proteasome beat subunit; K02732 20S proteasome s... 167 2e-41
xla:380127 psmb1, MGC154302, MGC52861; proteasome (prosome, ma... 164 2e-40
hsa:5689 PSMB1, FLJ25321, HC5, KIAA1838, PMSB1, PSC5; proteaso... 164 3e-40
mmu:19170 Psmb1, AA409053, C81484, Lmpc5; proteasome (prosome,... 162 7e-40
pfa:PFE0915c proteasome subunit beta type 1, putative (EC:3.4.... 160 2e-39
dre:445413 psmb1, MGC103665, wu:fc51f06, zgc:103665; proteasom... 159 6e-39
mmu:631331 Gm7061, EG631331; predicted gene 7061 129 7e-30
sce:YBL041W PRE7, PRS3; Beta 6 subunit of the 20S proteasome (... 125 2e-28
cel:C02F5.9 pbs-6; Proteasome Beta Subunit family member (pbs-... 123 4e-28
cpv:cgd2_530 possible proteasome component ; K02735 20S protea... 71.6 2e-12
tpv:TP04_0618 proteasome subunit beta type 3 (EC:3.4.25.1); K0... 70.1 6e-12
pfa:PFA_0400c beta3 proteasome subunit, putative (EC:3.4.25.1)... 62.0 1e-09
cel:Y38A8.2 pbs-3; Proteasome Beta Subunit family member (pbs-... 61.2 3e-09
xla:496005 psmb3, HC10-II; proteasome (prosome, macropain) sub... 57.4 4e-08
tgo:TGME49_114090 proteasome subunit beta type 3, putative (EC... 56.6 6e-08
ath:AT1G21720 PBC1; PBC1 (PROTEASOME BETA SUBUNIT C1); peptida... 56.6 7e-08
sce:YER094C PUP3, SCS32; Beta 3 subunit of the 20S proteasome ... 55.1 2e-07
ath:AT1G77440 PBC2; PBC2; peptidase/ threonine-type endopeptid... 55.1 2e-07
hsa:5691 PSMB3, HC10-II, MGC4147; proteasome (prosome, macropa... 55.1 2e-07
mmu:26446 Psmb3, AL033320, C10-II, MGC106850; proteasome (pros... 54.7 2e-07
tgo:TGME49_023590 proteasome component PRE3 precursor, putativ... 54.3 3e-07
cel:K08D12.1 pbs-1; Proteasome Beta Subunit family member (pbs... 54.3 3e-07
dre:573095 psmb3, MGC56374, wu:fb11d10, wu:fu88b08, zgc:56374;... 53.5 5e-07
bbo:BBOV_III007090 17.m07625; proteasome A-type and B-type fam... 52.4 1e-06
mmu:100505005 proteasome subunit beta type-3-like 45.8 1e-04
ath:AT3G26340 20S proteasome beta subunit E, putative; K02737 ... 43.9 4e-04
ath:AT4G31300 PBA1; PBA1; endopeptidase/ peptidase/ threonine-... 43.5 5e-04
ath:AT1G13060 PBE1; PBE1; endopeptidase/ peptidase/ threonine-... 43.1 6e-04
cpv:cgd2_860 Pre3p/proteasome regulatory subunit beta type 6, ... 43.1 6e-04
tgo:TGME49_106930 proteasome subunit beta type 7, putative (EC... 42.4 0.001
dre:30666 psmb8, lmp7, macropain, prosome; proteasome (prosome... 41.2 0.003
cpv:cgd3_2530 PUP1/proteasome subunit beta type 7, NTN hydrola... 41.2 0.003
dre:30665 psmb9a, lmp2, macropain, prosome, psmb9, wu:fa66f04,... 40.8 0.003
sce:YJL001W PRE3, CRL21; Beta 1 subunit of the 20S proteasome,... 40.8 0.004
xla:378667 psmb9, LMP2, psmb9-A; proteasome (prosome, macropai... 39.3 0.009
xla:431796 hypothetical protein MGC84496; K02734 20S proteasom... 38.5 0.018
pfa:PF10_0111 20S proteasome beta subunit, putative; K02737 20... 38.1 0.020
dre:436882 psmb2, zgc:92282; proteasome (prosome, macropain) s... 37.7 0.026
xla:443966 psmb2, MGC130720, MGC80364; proteasome (prosome, ma... 37.4 0.041
tpv:TP01_0908 proteasome subunit beta type 7 precursor; K02739... 37.0 0.045
hsa:122706 PSMB11, BETA5T, FLJ16369; proteasome (prosome, macr... 37.0 0.057
mmu:73902 Psmb11, 5830406J20Rik, beta5t; proteasome (prosome, ... 36.2 0.084
dre:446135 si:ch211-14a17.13 (EC:3.4.25.1); K11598 20S proteas... 35.8 0.10
dre:100331997 proteasome beta 11 subunit-like 35.8 0.10
dre:100331481 proteasome beta 5 subunit-like 35.4 0.14
hsa:5696 PSMB8, D6S216, D6S216E, LMP7, MGC1491, PSMB5i, RING10... 35.4 0.15
> tgo:TGME49_090010 proteasome subunit beta type 1, putative (EC:3.4.25.1);
K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=359
Score = 221 bits (563), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 106/171 (61%), Positives = 131/171 (76%), Gaps = 2/171 (1%)
Query 42 QQTALPLAEISSRHGRGGFNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKI 101
++ LP+A + R F+PYVNNGGTVVCVAGEDF + VGDTRLS G+SI+SR SKI
Sbjct 133 EEAILPVALAAPMERR--FSPYVNNGGTVVCVAGEDFAVAVGDTRLSTGFSIYSRRQSKI 190
Query 102 TQLTSRCCIASSGMQADINTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFP 161
T+LT + +A+SGM+AD TLH LLK R+ Y H HR PS A++QLLS++LYSRRFFP
Sbjct 191 TKLTDKVVLATSGMEADKTTLHNLLKIRIEQYTHQHRHPPSLNAIAQLLSTVLYSRRFFP 250
Query 162 FYTFNLLFALDEEGKGAVFGYDAIGSFERSSFNAAGTGGPLVTALLDNQIA 212
FYTFN+L +DE GKGAV+GYDAIGSFE S +N AGTG L+ +LDNQI+
Sbjct 251 FYTFNVLCGIDENGKGAVYGYDAIGSFEPSRYNCAGTGAHLIMPVLDNQIS 301
> cpv:cgd1_2490 proteasome subunit beta type 1 ; K02732 20S proteasome
subunit beta 6 [EC:3.4.25.1]
Length=246
Score = 188 bits (477), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 89/153 (58%), Positives = 114/153 (74%), Gaps = 0/153 (0%)
Query 60 FNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADI 119
FNPY+NNGG+ V VAG+DFV+ DTRLS Y I SR SK QLT++C +A SGM ADI
Sbjct 37 FNPYMNNGGSCVAVAGDDFVVIAADTRLSKMYRIASRSVSKTCQLTNKCILACSGMLADI 96
Query 120 NTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAV 179
N L K L A+V +Y H PS A++QLLS +LYS+RFFP+Y+F LL LDE+GKG V
Sbjct 97 NALRKTLLAKVKLYEFEHNKTPSINAIAQLLSCILYSKRFFPYYSFCLLSGLDEQGKGVV 156
Query 180 FGYDAIGSFERSSFNAAGTGGPLVTALLDNQIA 212
+GYDA+GSF++ F A G+GG L+T++LDNQI+
Sbjct 157 YGYDAVGSFDQHKFVALGSGGSLITSILDNQIS 189
> ath:AT3G60820 PBF1; PBF1; peptidase/ threonine-type endopeptidase;
K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=223
Score = 179 bits (455), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 81/154 (52%), Positives = 109/154 (70%), Gaps = 0/154 (0%)
Query 58 GGFNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQA 117
++PY NNGGT V +AG D+ + DTR+S GYSI SR +SKI +L R ++SSG QA
Sbjct 6 ANWSPYDNNGGTCVAIAGSDYCVIAADTRMSTGYSILSRDYSKIHKLADRAVLSSSGFQA 65
Query 118 DINTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKG 177
D+ L K+LK+R +Y+H H + S PA++QLLS+ LY +RFFP+Y FN+L LDEEGKG
Sbjct 66 DVKALQKVLKSRHLIYQHQHNKQMSCPAMAQLLSNTLYFKRFFPYYAFNVLGGLDEEGKG 125
Query 178 AVFGYDAIGSFERSSFNAAGTGGPLVTALLDNQI 211
VF YDA+GS+ER + A G+G L+ LDNQ+
Sbjct 126 CVFTYDAVGSYERVGYGAQGSGSTLIMPFLDNQL 159
> bbo:BBOV_II002710 18.m06221; proteasome A-type and B-type family
protein; K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=217
Score = 168 bits (426), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 75/152 (49%), Positives = 107/152 (70%), Gaps = 0/152 (0%)
Query 60 FNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADI 119
FNPYVNNGGTVV + + DTRLS+GY+IH+R+ SK+ +LT C I +SGMQAD+
Sbjct 7 FNPYVNNGGTVVAAVWNNCAVIAADTRLSIGYAIHTRNVSKLKKLTQTCVIGTSGMQADM 66
Query 120 NTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAV 179
+ LH L+ ++ +YR H EP ++QLLS++LYSRRFFP+YTFN+L +DE G+G
Sbjct 67 HALHSALERQIELYRFKHHREPGISVIAQLLSTILYSRRFFPYYTFNILCGVDENGRGIT 126
Query 180 FGYDAIGSFERSSFNAAGTGGPLVTALLDNQI 211
G+DAIG++ ++ A GT L+ ++LDN +
Sbjct 127 CGFDAIGNYGFDAYMAKGTSSSLIMSILDNHL 158
> tpv:TP04_0315 proteasome beat subunit; K02732 20S proteasome
subunit beta 6 [EC:3.4.25.1]
Length=218
Score = 167 bits (424), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 73/150 (48%), Positives = 106/150 (70%), Gaps = 0/150 (0%)
Query 60 FNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADI 119
FNPYVNNGGTV+ ++ + DTRLS Y IH+R +K+ +LT +C + +SGMQAD+
Sbjct 8 FNPYVNNGGTVIAATWNNYAVIAADTRLSQRYLIHTRFSTKLFKLTDKCILGTSGMQADM 67
Query 120 NTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAV 179
L +L+ ++ +YR+ H EP+ A++QLLS++LY RRFFP+YTFN+L +DE+GKG V
Sbjct 68 LALQSVLERQIELYRYTHNREPTFNAIAQLLSTVLYGRRFFPYYTFNILCGIDEQGKGVV 127
Query 180 FGYDAIGSFERSSFNAAGTGGPLVTALLDN 209
GYDA+G++ + A GT LV ++LDN
Sbjct 128 CGYDAVGNYNYEKYTAQGTSSSLVISVLDN 157
> xla:380127 psmb1, MGC154302, MGC52861; proteasome (prosome,
macropain) subunit, beta type, 1 (EC:3.4.25.1); K02732 20S proteasome
subunit beta 6 [EC:3.4.25.1]
Length=239
Score = 164 bits (415), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 73/152 (48%), Positives = 108/152 (71%), Gaps = 0/152 (0%)
Query 60 FNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADI 119
FNPY NGGTV+ +AG+DF + DTRLS GYSIHSR+ K +LT + I +G AD
Sbjct 28 FNPYTFNGGTVLALAGDDFALVASDTRLSEGYSIHSRNTPKCYKLTDKTVIGCTGFHADC 87
Query 120 NTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAV 179
TL K+++AR+ +Y+H + + A++ +LS++LYSRRFFP+Y +N++ LDEEGKGAV
Sbjct 88 LTLTKIIEARLKMYKHSNNKTMTSGAIAAMLSTILYSRRFFPYYVYNIIGGLDEEGKGAV 147
Query 180 FGYDAIGSFERSSFNAAGTGGPLVTALLDNQI 211
+ +D +GS++R ++ A G+ ++ LLDNQI
Sbjct 148 YSFDPVGSYQRDAYKAGGSASAMLQPLLDNQI 179
> hsa:5689 PSMB1, FLJ25321, HC5, KIAA1838, PMSB1, PSC5; proteasome
(prosome, macropain) subunit, beta type, 1 (EC:3.4.25.1);
K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=241
Score = 164 bits (414), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 73/152 (48%), Positives = 107/152 (70%), Gaps = 0/152 (0%)
Query 60 FNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADI 119
F+PYV NGGT++ +AGEDF I DTRLS G+SIH+R K +LT + I SG D
Sbjct 30 FSPYVFNGGTILAIAGEDFAIVASDTRLSEGFSIHTRDSPKCYKLTDKTVIGCSGFHGDC 89
Query 120 NTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAV 179
TL K+++AR+ +Y+H + + A++ +LS++LYSRRFFP+Y +N++ LDEEGKGAV
Sbjct 90 LTLTKIIEARLKMYKHSNNKAMTTGAIAAMLSTILYSRRFFPYYVYNIIGGLDEEGKGAV 149
Query 180 FGYDAIGSFERSSFNAAGTGGPLVTALLDNQI 211
+ +D +GS++R SF A G+ ++ LLDNQ+
Sbjct 150 YSFDPVGSYQRDSFKAGGSASAMLQPLLDNQV 181
> mmu:19170 Psmb1, AA409053, C81484, Lmpc5; proteasome (prosome,
macropain) subunit, beta type 1 (EC:3.4.25.1); K02732 20S
proteasome subunit beta 6 [EC:3.4.25.1]
Length=240
Score = 162 bits (410), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 76/162 (46%), Positives = 109/162 (67%), Gaps = 5/162 (3%)
Query 55 HGRGG-----FNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCC 109
HG G F+PY NGGTV+ +AGEDF I DTRLS G+SIH+R K +LT +
Sbjct 19 HGSAGPVQLRFSPYAFNGGTVLAIAGEDFSIVASDTRLSEGFSIHTRDSPKCYKLTDKTV 78
Query 110 IASSGMQADINTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLF 169
I SG D TL K+++AR+ +Y+H + + A++ +LS++LYSRRFFP+Y +N++
Sbjct 79 IGCSGFHGDCLTLTKIIEARLKMYKHSNNKAMTTGAIAAMLSTILYSRRFFPYYVYNIIG 138
Query 170 ALDEEGKGAVFGYDAIGSFERSSFNAAGTGGPLVTALLDNQI 211
LDEEGKGAV+ +D +GS++R SF A G+ ++ LLDNQ+
Sbjct 139 GLDEEGKGAVYSFDPVGSYQRDSFKAGGSASAMLQPLLDNQV 180
> pfa:PFE0915c proteasome subunit beta type 1, putative (EC:3.4.25.1);
K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=240
Score = 160 bits (406), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 72/160 (45%), Positives = 111/160 (69%), Gaps = 2/160 (1%)
Query 54 RHGRG--GFNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIA 111
HGRG + PY++NGGTV+ + G+D+VI DTRLS+ YSI++R KI++LT +C I
Sbjct 22 EHGRGFKRWYPYIDNGGTVIGLTGKDYVILAADTRLSLSYSIYTRFCPKISKLTDKCIIG 81
Query 112 SSGMQADINTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFAL 171
SSGMQ+DI TLH LL+ ++ ++ H P +++LL +LYSRRFFP+Y FN+L +
Sbjct 82 SSGMQSDIKTLHSLLQKKIQLFVLEHSHYPDIHVIARLLCVILYSRRFFPYYAFNILAGV 141
Query 172 DEEGKGAVFGYDAIGSFERSSFNAAGTGGPLVTALLDNQI 211
DE KG ++ YD++GS+ ++ + G+G L+ +LDN++
Sbjct 142 DENNKGVLYNYDSVGSYCEATHSCVGSGSQLILPILDNRV 181
> dre:445413 psmb1, MGC103665, wu:fc51f06, zgc:103665; proteasome
(prosome, macropain) subunit, beta type, 1 (EC:3.4.25.1);
K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=237
Score = 159 bits (402), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 72/153 (47%), Positives = 104/153 (67%), Gaps = 0/153 (0%)
Query 60 FNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADI 119
F+PY NGGTV+ VAGEDF I DTRLS GYSIHSR K +LT + SG D
Sbjct 26 FSPYAFNGGTVLAVAGEDFAIVASDTRLSEGYSIHSRDSPKCYKLTDTTVLGCSGFHGDC 85
Query 120 NTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAV 179
TL K+++AR+ +Y+H + + A++ +LS++LY RRFFP+Y +N++ LDEEG+GAV
Sbjct 86 LTLTKIIEARLKMYKHSNNKSMTSGAIAAMLSTILYGRRFFPYYVYNIIGGLDEEGRGAV 145
Query 180 FGYDAIGSFERSSFNAAGTGGPLVTALLDNQIA 212
+ +D +GS++R ++ A G+ ++ LLDNQI
Sbjct 146 YSFDPVGSYQRDTYKAGGSASAMLQPLLDNQIG 178
> mmu:631331 Gm7061, EG631331; predicted gene 7061
Length=730
Score = 129 bits (324), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 65/151 (43%), Positives = 96/151 (63%), Gaps = 5/151 (3%)
Query 60 FNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADI 119
F+PY NGG V+ +AGEDF I DTRLS +SIH+R K +LT + I SG D
Sbjct 29 FSPYAFNGGPVLAIAGEDFSIVASDTRLSEEFSIHTRDSPKCYKLTDKTVIGCSGFHGDC 88
Query 120 NTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAV 179
TL K+++ R+ +++A +G A++ +LS++ SRRFFP+Y +N++ LDEEGKGAV
Sbjct 89 LTLTKIIETRL----KMNKAMRTG-AVAAMLSTIPCSRRFFPYYVYNIIGGLDEEGKGAV 143
Query 180 FGYDAIGSFERSSFNAAGTGGPLVTALLDNQ 210
+ +D +GS++R S A + + LL NQ
Sbjct 144 YSFDPVGSYQRDSVKAGASASATLQPLLHNQ 174
> sce:YBL041W PRE7, PRS3; Beta 6 subunit of the 20S proteasome
(EC:3.4.25.1); K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=241
Score = 125 bits (313), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 61/153 (39%), Positives = 90/153 (58%), Gaps = 1/153 (0%)
Query 60 FNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADI 119
FNPY +NGGT++ +AGEDF + GDTR YSI+SR+ K+ ++++G AD
Sbjct 21 FNPYGDNGGTILGIAGEDFAVLAGDTRNITDYSINSRYEPKVFDCGDNIVMSANGFAADG 80
Query 120 NTLHKLLKARVAVYRHLHRAEP-SGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGA 178
+ L K K V Y H + S + ++ + LLY +RFFP+Y ++ LDE+GKGA
Sbjct 81 DALVKRFKNSVKWYHFDHNDKKLSINSAARNIQHLLYGKRFFPYYVHTIIAGLDEDGKGA 140
Query 179 VFGYDAIGSFERSSFNAAGTGGPLVTALLDNQI 211
V+ +D +GS+ER A G L+ LDNQ+
Sbjct 141 VYSFDPVGSYEREQCRAGGAAASLIMPFLDNQV 173
> cel:C02F5.9 pbs-6; Proteasome Beta Subunit family member (pbs-6);
K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=258
Score = 123 bits (309), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 68/174 (39%), Positives = 100/174 (57%), Gaps = 2/174 (1%)
Query 40 AAQQTALPLAEISSRH-GRGGFNPYVNNGGTVVCVAGEDFVIGVGDTRLSVG-YSIHSRH 97
A ++ A +SSR R +NPY GG+ ++GE+F I DTR++ +I +R
Sbjct 23 AMKEVAAHPEWMSSRQIERQRWNPYSMEGGSTCAISGENFAIVASDTRMTQNDINILTRD 82
Query 98 HSKITQLTSRCCIASSGMQADINTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSR 157
KI L + +SG D+ L K+L++R+ YR +R++ S ++LLS LY R
Sbjct 83 AEKIQILNDNIILTTSGFYGDVLQLKKVLQSRLHKYRFDYRSDMSVDLCAELLSRNLYYR 142
Query 158 RFFPFYTFNLLFALDEEGKGAVFGYDAIGSFERSSFNAAGTGGPLVTALLDNQI 211
RFFP+YT +L +DE GKGAVF YD IG ER ++A+G P++ LD QI
Sbjct 143 RFFPYYTGAILAGIDEHGKGAVFSYDPIGCIERLGYSASGAAEPMIIPFLDCQI 196
> cpv:cgd2_530 possible proteasome component ; K02735 20S proteasome
subunit beta 3 [EC:3.4.25.1]
Length=204
Score = 71.6 bits (174), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 48/148 (32%), Positives = 74/148 (50%), Gaps = 10/148 (6%)
Query 66 NGGTVVCVAGEDFVIGVGDTRL-SVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHK 124
NGG ++ + G++ V D RL S GY S K+ + +S+ + SG+ DI+TL
Sbjct 6 NGGAILAMKGKECVAFATDMRLGSNGYRTISTDFDKVIRPSSKTLMGFSGLATDIHTLTN 65
Query 125 LLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVFGYDA 184
L+K + +Y E ALS + +S+LYS+RF P++ ++ LD + GYD
Sbjct 66 LIKFKTNLYHLREEREIGVKALSHMTASILYSKRFSPYFVEPIVAGLDNNNVPFIAGYDL 125
Query 185 IGSFER-SSFNAAGTGGPLVTALLDNQI 211
IG S F +GT DNQ+
Sbjct 126 IGCLSVCSEFAISGTA--------DNQL 145
> tpv:TP04_0618 proteasome subunit beta type 3 (EC:3.4.25.1);
K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=206
Score = 70.1 bits (170), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 43/135 (31%), Positives = 71/135 (52%), Gaps = 2/135 (1%)
Query 66 NGGTVVCVAGEDFVIGVGDTRLSVGYSIH-SRHHSKITQLTSRCCIASSGMQADINTLHK 124
NGG VV + GE V D RL + + S + K ++T C A+SG+ D+ TL +
Sbjct 8 NGGAVVAMMGEGCVAIACDKRLGLNQQVTVSSNFPKAFKVTESCFFAASGLATDVQTLKE 67
Query 125 LLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVFGYDA 184
+ +V +Y+ E S LS ++ ++LYSRRF P++ +++ LD + + +D
Sbjct 68 EIMFKVNMYKLRGDKEMSVKTLSNMVGAMLYSRRFGPWFVDSVIAGLDNDSSPYITCFDL 127
Query 185 IGS-FERSSFNAAGT 198
+G+ S F AGT
Sbjct 128 VGAPCTPSDFVVAGT 142
> pfa:PFA_0400c beta3 proteasome subunit, putative (EC:3.4.25.1);
K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=204
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 41/128 (32%), Positives = 64/128 (50%), Gaps = 13/128 (10%)
Query 73 VAGEDFVIGVGDTRLSVG-YSIHSRHHSKITQLTSRCCIASSGMQADINTLHKLLKARVA 131
++G + V D RL ++ S SKI ++ + + SG+ DI TL+++L+ RV
Sbjct 1 MSGSNCVAIACDLRLGANTFTTVSTKFSKIFKMNNNVYVGLSGLATDIQTLYEILRYRVN 60
Query 132 VYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYT------FNLLFALDEEGKGAV------ 179
+Y AE + +LSS+LYS RF P++ F L +DEEG+ V
Sbjct 61 LYEVRQDAEMDVECFANMLSSILYSNRFSPYFVNPIVVGFKLKHYVDEEGEKKVNYEPYL 120
Query 180 FGYDAIGS 187
YD IG+
Sbjct 121 TAYDLIGA 128
> cel:Y38A8.2 pbs-3; Proteasome Beta Subunit family member (pbs-3);
K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=204
Score = 61.2 bits (147), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 39/144 (27%), Positives = 68/144 (47%), Gaps = 1/144 (0%)
Query 67 GGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKLL 126
GGTVV +AG++ V D R+ + + K+ ++T + + +G Q+D T+ + +
Sbjct 8 GGTVVAMAGDECVCIASDLRIGEQMTTIATDQKKVHKVTDKVYVGLAGFQSDARTVLEKI 67
Query 127 KARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVFGYDAIG 186
R +Y LS+++S+L Y RF ++T L+ LD+ K + D IG
Sbjct 68 MFRKNLYELRENRNIKPQVLSEMISNLAYQHRFGSYFTEPLVAGLDDTNKPYICCMDTIG 127
Query 187 SFER-SSFNAAGTGGPLVTALLDN 209
F A GTG + + +N
Sbjct 128 CVSAPRDFVAVGTGQEYLLGVCEN 151
> xla:496005 psmb3, HC10-II; proteasome (prosome, macropain) subunit,
beta type, 3; K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=205
Score = 57.4 bits (137), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 37/135 (27%), Positives = 63/135 (46%), Gaps = 2/135 (1%)
Query 66 NGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKL 125
NGG ++ + G+D V D R V + + KI + R I +G+ D+ T+ +
Sbjct 7 NGGAIMAMKGKDCVAIAADRRFGVQAQMVTTDFQKIFPMGERLYIGLAGLATDVQTVAQR 66
Query 126 LKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEG-KGAVFGYDA 184
LK R+ +Y + ++++LLY RRF P+Y ++ LD + + + D
Sbjct 67 LKFRLNLYELKEGRQIKPKTFMSMVANLLYERRFGPYYIEPVIAGLDPKTFQPFICSLDL 126
Query 185 IGS-FERSSFNAAGT 198
IG E F +GT
Sbjct 127 IGCPMETEDFVVSGT 141
> tgo:TGME49_114090 proteasome subunit beta type 3, putative (EC:3.4.25.1);
K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=205
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 38/125 (30%), Positives = 62/125 (49%), Gaps = 3/125 (2%)
Query 66 NGGTVVCVAGEDFVIGVGDTRLSVG-YSIHSRHHSKITQLTSRCCIASSGMQADINTLHK 124
NG VV +AG+D V DTRL V + S K+ ++ + + +G+ D+ T+HK
Sbjct 5 NGSAVVAMAGKDCVGIASDTRLGVNQFGTVSADFQKVFKMNNHTFVGLAGLATDVQTVHK 64
Query 125 LLKARVAVYRHLHRAEPSGPA-LSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVF-GY 182
L R +Y+ P +S ++SS+LY RRF P++ ++ L E +
Sbjct 65 ELVFRSNLYQLREEVTEMPPEIMSNVVSSMLYGRRFAPYFVSPVVAGLHPETHQPFLSAF 124
Query 183 DAIGS 187
D IG+
Sbjct 125 DYIGA 129
> ath:AT1G21720 PBC1; PBC1 (PROTEASOME BETA SUBUNIT C1); peptidase/
threonine-type endopeptidase; K02735 20S proteasome subunit
beta 3 [EC:3.4.25.1]
Length=204
Score = 56.6 bits (135), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 36/135 (26%), Positives = 66/135 (48%), Gaps = 1/135 (0%)
Query 66 NGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKL 125
NG VV + G++ D RL V + +I+++ R I SG+ D+ TL++
Sbjct 7 NGSAVVAMVGKNCFAIASDRRLGVQLQTIATDFQRISKIHDRVFIGLSGLATDVQTLYQR 66
Query 126 LKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVFGYDAI 185
L R +Y+ + + L+S++LY +RF P+ ++ L ++ K + D+I
Sbjct 67 LVFRHKLYQLREERDMKPETFASLVSAILYEKRFGPYLCQPVIAGLGDDDKPFICTMDSI 126
Query 186 GSFERS-SFNAAGTG 199
G+ E + F +GT
Sbjct 127 GAKELAKDFVVSGTA 141
> sce:YER094C PUP3, SCS32; Beta 3 subunit of the 20S proteasome
involved in ubiquitin-dependent catabolism; human homolog
is subunit C10 (EC:3.4.25.1); K02735 20S proteasome subunit
beta 3 [EC:3.4.25.1]
Length=205
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 40/140 (28%), Positives = 66/140 (47%), Gaps = 3/140 (2%)
Query 61 NPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADIN 120
+P NGG VV + G+D V D RL S KI + +G+ D+
Sbjct 3 DPSSINGGIVVAMTGKDCVAIACDLRLGSQSLGVSNKFEKIFHY-GHVFLGITGLATDVT 61
Query 121 TLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALD-EEGKGAV 179
TL+++ + + +Y+ +QL+SS LY RRF P++ ++ ++ + GK +
Sbjct 62 TLNEMFRYKTNLYKLKEERAIEPETFTQLVSSSLYERRFGPYFVGPVVAGINSKSGKPFI 121
Query 180 FGYDAIGSF-ERSSFNAAGT 198
G+D IG E F +GT
Sbjct 122 AGFDLIGCIDEAKDFIVSGT 141
> ath:AT1G77440 PBC2; PBC2; peptidase/ threonine-type endopeptidase;
K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=204
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/135 (26%), Positives = 65/135 (48%), Gaps = 1/135 (0%)
Query 66 NGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKL 125
NG VV + G++ D RL V + +I+++ I SG+ D+ TL++
Sbjct 7 NGSAVVAMVGKNCFAIASDRRLGVQLQTIATDFQRISKIHDHLFIGLSGLATDVQTLYQR 66
Query 126 LKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVFGYDAI 185
L R +Y+ + + L+S++LY +RF PF ++ L ++ K + D+I
Sbjct 67 LVFRHKLYQLREERDMKPETFASLVSAILYEKRFGPFLCQPVIAGLGDDNKPFICTMDSI 126
Query 186 GSFERS-SFNAAGTG 199
G+ E + F +GT
Sbjct 127 GAKELAKDFVVSGTA 141
> hsa:5691 PSMB3, HC10-II, MGC4147; proteasome (prosome, macropain)
subunit, beta type, 3 (EC:3.4.25.1); K02735 20S proteasome
subunit beta 3 [EC:3.4.25.1]
Length=205
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 37/135 (27%), Positives = 64/135 (47%), Gaps = 2/135 (1%)
Query 66 NGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKL 125
NGG V+ + G++ V D R + + + KI + R I +G+ D+ T+ +
Sbjct 7 NGGAVMAMKGKNCVAIAADRRFGIQAQMVTTDFQKIFPMGDRLYIGLAGLATDVQTVAQR 66
Query 126 LKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEG-KGAVFGYDA 184
LK R+ +Y + L ++++LLY +RF P+YT ++ LD + K + D
Sbjct 67 LKFRLNLYELKEGRQIKPYTLMSMVANLLYEKRFGPYYTEPVIAGLDPKTFKPFICSLDL 126
Query 185 IGS-FERSSFNAAGT 198
IG F +GT
Sbjct 127 IGCPMVTDDFVVSGT 141
> mmu:26446 Psmb3, AL033320, C10-II, MGC106850; proteasome (prosome,
macropain) subunit, beta type 3 (EC:3.4.25.1); K02735
20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=205
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 37/135 (27%), Positives = 64/135 (47%), Gaps = 2/135 (1%)
Query 66 NGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKL 125
NGG V+ + G++ V D R + + + KI + R I +G+ D+ T+ +
Sbjct 7 NGGAVMAMKGKNCVAIAADRRFGIQAQMVTTDFQKIFPMGDRLYIGLAGLATDVQTVAQR 66
Query 126 LKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEG-KGAVFGYDA 184
LK R+ +Y + L ++++LLY +RF P+YT ++ LD + K + D
Sbjct 67 LKFRLNLYELKEGRQIKPYTLMSMVANLLYEKRFGPYYTEPVIAGLDPKTFKPFICSLDL 126
Query 185 IGS-FERSSFNAAGT 198
IG F +GT
Sbjct 127 IGCPMVTDDFVVSGT 141
> tgo:TGME49_023590 proteasome component PRE3 precursor, putative
(EC:3.4.25.1); K02738 20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=245
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 47/172 (27%), Positives = 82/172 (47%), Gaps = 8/172 (4%)
Query 40 AAQQTALPLAEISSRHGRGGFNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHS 99
A+ LP+ +S+ GRG V+ G T+V V+ + V+ DTR S G + +R
Sbjct 21 ASSPAGLPVPSLSA-SGRG--PTAVSTGTTIVAVSFKGGVVLGADTRTSAGSYVVNRAAR 77
Query 100 KITQLTSRCCIASSGMQADINTLHKLLKARVAVY-RHLHRA-EPSGPALSQLLSSLLYSR 157
KI+++ R C+ SG AD + +++K + Y + L + EP A + + SL Y
Sbjct 78 KISRVHERICVCRSGSAADTQAVTQIVKLYIQQYAQELPKGEEPRVEAAANVFQSLCYQH 137
Query 158 RFFPFYTFNLLFA-LDEEGKGAVFGYDAIGSFERSSFNAAGTGGPLVTALLD 208
+ T L+ A D+ G ++ G+ + A G+G ++A +D
Sbjct 138 K--DALTAGLIVAGFDKVKGGQIYALPLGGALVPMQYTAGGSGSAFISAYMD 187
> cel:K08D12.1 pbs-1; Proteasome Beta Subunit family member (pbs-1);
K02738 20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=239
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 44/154 (28%), Positives = 65/154 (42%), Gaps = 3/154 (1%)
Query 59 GFNPY--VNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQ 116
GF P ++ G T++ + V+ D+R S G I SR +KIT +T + SG
Sbjct 13 GFYPQEEISTGTTLIAMEYNGGVVVGTDSRTSAGSFITSRATNKITPITDNMVVCRSGSA 72
Query 117 ADINTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGK 176
AD + + K + VY + SQ+ LY+ R L+ DEE
Sbjct 73 ADTQAIADIAKYHIDVYTMTENKPVTIYRSSQIFRQFLYNYR-EQLSASVLVAGWDEELG 131
Query 177 GAVFGYDAIGSFERSSFNAAGTGGPLVTALLDNQ 210
G V+ G R A+G+G V LD+Q
Sbjct 132 GQVYAIPIGGFVSRQRSTASGSGSTFVQGFLDSQ 165
> dre:573095 psmb3, MGC56374, wu:fb11d10, wu:fu88b08, zgc:56374;
proteasome (prosome, macropain) subunit, beta type, 3 (EC:3.4.25.1);
K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=205
Score = 53.5 bits (127), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 52/107 (48%), Gaps = 0/107 (0%)
Query 66 NGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKL 125
NGG V+ + G++ V D R + + + KI + R I +G+ D+ T+ +
Sbjct 7 NGGAVMAMRGKECVAIASDRRFGIQAQLVTTDFQKIFPMGERLYIGLAGLATDVQTVSQR 66
Query 126 LKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALD 172
LK R+ +Y + ++S+LLY RRF P+Y ++ LD
Sbjct 67 LKFRLNLYELKEGRQIKPRTFMSMVSNLLYERRFGPYYIEPVIAGLD 113
> bbo:BBOV_III007090 17.m07625; proteasome A-type and B-type family
protein; K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=205
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 35/123 (28%), Positives = 61/123 (49%), Gaps = 2/123 (1%)
Query 66 NGGTVVCVAGEDFVIGVGDTRLSV-GYSIHSRHHSKITQLTSRCCIASSGMQADINTLHK 124
NGG VV + G V D RL + G S + K ++T A+ G+ DI T+
Sbjct 8 NGGAVVAMVGNGCVAIACDKRLGMNGQHTISSNFPKAFKVTDTAYFAACGLATDIQTMKS 67
Query 125 LLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVFGYDA 184
++ + +Y L+ ++ SLLYSRRF P++ +++ LD++ ++ +D
Sbjct 68 EIEFKTNMYALRCEKNMGVKTLAHMVGSLLYSRRFGPWFVSSVVAGLDKDTP-HIYCFDL 126
Query 185 IGS 187
IG+
Sbjct 127 IGA 129
> mmu:100505005 proteasome subunit beta type-3-like
Length=205
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/135 (24%), Positives = 61/135 (45%), Gaps = 2/135 (1%)
Query 66 NGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKL 125
NGG V+ + ++ V D R + + + KI + R + +G+ D+ T+ +
Sbjct 7 NGGAVMAMKEKNCVAIAADRRFGIQAQMVTTDFQKIFPMGDRLYVGLAGLAIDVQTVAQR 66
Query 126 LKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEG-KGAVFGYDA 184
LK + +Y + L ++++LLY + F P+YT ++ LD + K + D
Sbjct 67 LKFLLNLYELKEGWQIKPYTLMSMVANLLYEKWFGPYYTEPVIAGLDPKTFKPFICSLDL 126
Query 185 IGS-FERSSFNAAGT 198
IG F +GT
Sbjct 127 IGCPMVTDDFVVSGT 141
> ath:AT3G26340 20S proteasome beta subunit E, putative; K02737
20S proteasome subunit beta 5 [EC:3.4.25.1]
Length=273
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 35/144 (24%), Positives = 64/144 (44%), Gaps = 1/144 (0%)
Query 66 NGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKL 125
G T + ++ V+ D+R S+G I S+ KI ++ +G AD H+
Sbjct 56 KGTTTLAFIFKEGVMVAADSRASMGGYISSQSVKKIIEINPYMLGTMAGGAADCQFWHRN 115
Query 126 LKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVFGYDAI 185
L + ++ ++ S S+LL+++LYS R ++ DE G G + +
Sbjct 116 LGIKCRLHELANKRRISVSGASKLLANMLYSYRGMGLSVGTMIAGWDETGPGLYYVDNEG 175
Query 186 GSFERSSFNAAGTGGPLVTALLDN 209
G + F + G+G P +LD+
Sbjct 176 GRLKGDRF-SVGSGSPYAYGVLDS 198
> ath:AT4G31300 PBA1; PBA1; endopeptidase/ peptidase/ threonine-type
endopeptidase; K02738 20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=234
Score = 43.5 bits (101), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 36/146 (24%), Positives = 60/146 (41%), Gaps = 8/146 (5%)
Query 67 GGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKLL 126
G T++ V V+ D+R S G + +R KITQLT + SG AD +++
Sbjct 12 GTTIIGVTYNGGVVLGADSRTSTGMYVANRASDKITQLTDNVYVCRSGSAAD----SQVV 67
Query 127 KARVAVYRHLHRAEPSGPAL----SQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVFGY 182
V + H H + PA + L+ L Y+ + T ++ D+ G ++G
Sbjct 68 SDYVRYFLHQHTIQHGQPATVKVSANLIRMLAYNNKQNMLQTGLIVGGWDKYEGGKIYGI 127
Query 183 DAIGSFERSSFNAAGTGGPLVTALLD 208
G+ F G+G + D
Sbjct 128 PLGGTVVEQPFAIGGSGSSYLYGFFD 153
> ath:AT1G13060 PBE1; PBE1; endopeptidase/ peptidase/ threonine-type
endopeptidase; K02737 20S proteasome subunit beta 5 [EC:3.4.25.1]
Length=274
Score = 43.1 bits (100), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 33/131 (25%), Positives = 59/131 (45%), Gaps = 1/131 (0%)
Query 79 VIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKLLKARVAVYRHLHR 138
V+ D+R S+G I S+ KI ++ +G AD H+ L + ++ ++
Sbjct 69 VMVAADSRASMGGYISSQSVKKIIEINPYMLGTMAGGAADCQFWHRNLGIKCRLHELANK 128
Query 139 AEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVFGYDAIGSFERSSFNAAGT 198
S S+LL+++LYS R ++ DE G G + + G + F + G+
Sbjct 129 RRISVSGASKLLANMLYSYRGMGLSVGTMIAGWDETGPGLYYVDNEGGRLKGDRF-SVGS 187
Query 199 GGPLVTALLDN 209
G P +LD+
Sbjct 188 GSPYAYGVLDS 198
> cpv:cgd2_860 Pre3p/proteasome regulatory subunit beta type 6,
NTN hydrolase fold ; K02738 20S proteasome subunit beta 1
[EC:3.4.25.1]
Length=248
Score = 43.1 bits (100), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 34/151 (22%), Positives = 66/151 (43%), Gaps = 1/151 (0%)
Query 60 FNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADI 119
N + G T+V + +D ++ D R S G I R KITQ+T + + SG AD
Sbjct 44 LNGEIETGTTIVALKYKDGLVLAADGRTSTGPIIAFRAARKITQITDKVFMCRSGSAADT 103
Query 120 NTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAV 179
+ + ++ R+ L E + ++ L+ + ++ +D G+ V
Sbjct 104 QIISRYVR-RIVQDHELETGEDTKVKSVASVARLISYQNKEHLLADMIIAGMDPNGEFKV 162
Query 180 FGYDAIGSFERSSFNAAGTGGPLVTALLDNQ 210
F G+ S+ +G+G + ++LD++
Sbjct 163 FRIPLGGTLIEGSYAISGSGSGYIYSMLDSK 193
> tgo:TGME49_106930 proteasome subunit beta type 7, putative (EC:3.4.25.1);
K02739 20S proteasome subunit beta 2 [EC:3.4.25.1]
Length=368
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 37/150 (24%), Positives = 69/150 (46%), Gaps = 2/150 (1%)
Query 59 GFNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQAD 118
G P G T+ V +D V+ DTR + G + ++ SK+ ++ A +G AD
Sbjct 126 GLPPARKTGTTICGVVCKDGVVLGADTRATEGTIVADKNCSKLHRIADNMYAAGAGTSAD 185
Query 119 INTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGA 178
++ + L +V ++R A+P +LS L+ + + +L +D +G
Sbjct 186 LDHMCDWLAVQVELHRLNTNAKPRVSMAVSVLSQELFKYQGYKGCAV-VLGGVDFKGP-Q 243
Query 179 VFGYDAIGSFERSSFNAAGTGGPLVTALLD 208
++ GS + S+F A G+G A+L+
Sbjct 244 IYKIHPHGSTDCSNFAAMGSGSLNAMAVLE 273
> dre:30666 psmb8, lmp7, macropain, prosome; proteasome (prosome,
macropain) subunit, beta type, 8 (EC:3.4.25.1); K02740 20S
proteasome subunit beta 8 [EC:3.4.25.1]
Length=271
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/117 (24%), Positives = 53/117 (45%), Gaps = 0/117 (0%)
Query 64 VNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLH 123
+N+G T + VI D+R S G I S+ +K+ ++ SG AD
Sbjct 64 LNHGTTTLAFKFRHGVIVAVDSRASAGKYIDSKEANKVIEINPYLLGTMSGSAADCQYWE 123
Query 124 KLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVF 180
+LL +Y+ ++ S A S+LLS+++ R +++ D++G G +
Sbjct 124 RLLAKECRLYKLRNKQRISVSAASKLLSNMMLGYRGMGLSMGSMICGWDKQGPGLYY 180
> cpv:cgd3_2530 PUP1/proteasome subunit beta type 7, NTN hydrolase
fold ; K02739 20S proteasome subunit beta 2 [EC:3.4.25.1]
Length=279
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 35/145 (24%), Positives = 65/145 (44%), Gaps = 2/145 (1%)
Query 64 VNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLH 123
V G T+V VA D V+ DTR + G + + KI +L+ A +G AD++ +
Sbjct 38 VKTGTTIVGVACNDCVVLGADTRATNGPIVADKDCEKIHRLSDNIFAAGAGTAADLDHVT 97
Query 124 KLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVFGYD 183
L++ + + + +P +LS LY ++ + +L+ A + VF
Sbjct 98 SLIEGNLELQKLQMNRKPRVAHAVSMLSDHLY--KYQGYIGAHLIVAGSDSTGNFVFQVS 155
Query 184 AIGSFERSSFNAAGTGGPLVTALLD 208
A G + F + G+G ++L+
Sbjct 156 ANGCIMQLPFTSMGSGSLCARSILE 180
> dre:30665 psmb9a, lmp2, macropain, prosome, psmb9, wu:fa66f04,
wu:fj22d05, zgc:91842; proteasome (prosome, macropain) subunit,
beta type, 9a (EC:3.4.25.1); K02741 20S proteasome subunit
beta 9 [EC:3.4.25.1]
Length=218
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 35/154 (22%), Positives = 75/154 (48%), Gaps = 4/154 (2%)
Query 58 GGFNPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQA 117
G + V G T++ V + V+ D+R+S G S+ +R +K++ L + A SG A
Sbjct 10 GWLSEEVKTGTTIIAVTFDGGVVIGSDSRVSAGESVVNRVMNKLSPLHDKIYCALSGSAA 69
Query 118 DINTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFA-LDEEGK 176
D T+ +++ ++ V+ +P + + L+ ++ Y ++ + +L+ A D++G
Sbjct 70 DAQTIAEIVNYQLDVHSIEVEDDPLVCSAATLVKNISY--KYKEELSAHLIVAGWDKKGG 127
Query 177 GAVFGYDAIGSFERSSFNAAGTGGPLVTALLDNQ 210
G V+ + G + F G+G + +D +
Sbjct 128 GQVYATLS-GLLTKQPFAIGGSGSFYINGFVDAE 160
> sce:YJL001W PRE3, CRL21; Beta 1 subunit of the 20S proteasome,
responsible for cleavage after acidic residues in peptides
(EC:3.4.25.1); K02738 20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=215
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 34/148 (22%), Positives = 65/148 (43%), Gaps = 4/148 (2%)
Query 64 VNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLH 123
V+ G +++ V +D VI D+R + G I +R K+T++ + SG AD +
Sbjct 16 VSLGTSIMAVTFKDGVILGADSRTTTGAYIANRVTDKLTRVHDKIWCCRSGSAADTQAIA 75
Query 124 KLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFA-LDEEGKGAVFGY 182
+++ + +Y + PS + + L Y + T ++ A D++ KG V+
Sbjct 76 DIVQYHLELYTSQY-GTPSTETAASVFKELCYENK--DNLTAGIIVAGYDDKNKGEVYTI 132
Query 183 DAIGSFERSSFNAAGTGGPLVTALLDNQ 210
GS + + AG+G + D
Sbjct 133 PLGGSVHKLPYAIAGSGSTFIYGYCDKN 160
> xla:378667 psmb9, LMP2, psmb9-A; proteasome (prosome, macropain)
subunit, beta type, 9 (large multifunctional peptidase
2); K02741 20S proteasome subunit beta 9 [EC:3.4.25.1]
Length=215
Score = 39.3 bits (90), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 38/155 (24%), Positives = 69/155 (44%), Gaps = 8/155 (5%)
Query 61 NPY----VNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQ 116
NPY V+ G T++ V + V+ D+R+S G ++ +R +K+ + R A SG
Sbjct 6 NPYTAREVSTGTTIIAVEFDGGVVLGSDSRVSAGDAVVNRVFNKLAPVHQRIYCALSGSA 65
Query 117 ADINTLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFA-LDEEG 175
AD + + + V+ A P A + L+ + Y ++ +L+ A D +
Sbjct 66 ADAQAVADMAHYHMEVHSIEMEAPPLVLAAANLIKGISY--KYKEEIMAHLIVAGWDRKH 123
Query 176 KGAVFGYDAIGSFERSSFNAAGTGGPLVTALLDNQ 210
G V+G G R F G+G + +D++
Sbjct 124 GGQVYGTLG-GMIIRQPFTIGGSGSTYIYGFVDSK 157
> xla:431796 hypothetical protein MGC84496; K02734 20S proteasome
subunit beta 4 [EC:3.4.25.1]
Length=199
Score = 38.5 bits (88), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 32/142 (22%), Positives = 62/142 (43%), Gaps = 4/142 (2%)
Query 70 VVCVAGEDFVIGVGDTRLSVGYSIHSRHH-SKITQLTSRCCIASSGMQADINTLHKLLKA 128
++ + G DFV+ DT + I +H K+ +++ + + G D + ++
Sbjct 4 LIGIQGNDFVLVAADT-VCANSIIQMKHDMDKMFKMSEKILLLCVGEAGDTVQFAEYIQK 62
Query 129 RVAVYRHLHRAEPSGPALSQLLSSLL--YSRRFFPFYTFNLLFALDEEGKGAVFGYDAIG 186
V +Y+ + E S A + L Y R P++ LL DE +++ D +
Sbjct 63 NVQLYKMRNGYELSPTAAANFTRRNLADYLRSRTPYHVNLLLAGYDEHAGPSLYYMDYLS 122
Query 187 SFERSSFNAAGTGGPLVTALLD 208
+ ++ F A G G L ++LD
Sbjct 123 ALAKTRFAAHGYGAYLTLSILD 144
> pfa:PF10_0111 20S proteasome beta subunit, putative; K02737
20S proteasome subunit beta 5 [EC:3.4.25.1]
Length=271
Score = 38.1 bits (87), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 62/142 (43%), Gaps = 1/142 (0%)
Query 67 GGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKLL 126
G T + +D +I D+R S+G I S++ KI ++ +G AD K L
Sbjct 60 GTTTLAFKFKDGIIVAVDSRASMGSFISSQNVEKIIEINKNILGTMAGGAADCLYWEKYL 119
Query 127 KARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVFGYDAIG 186
+ +Y + + S A S +LS++LY + + +L D G + D+
Sbjct 120 GKIIKIYELRNNEKISVRAASTILSNILYQYKGYGLCCGIILSGYDHTGFNMFYVDDSGK 179
Query 187 SFERSSFNAAGTGGPLVTALLD 208
E + F + G+G ++LD
Sbjct 180 KVEGNLF-SCGSGSTYAYSILD 200
> dre:436882 psmb2, zgc:92282; proteasome (prosome, macropain)
subunit, beta type, 2 (EC:3.4.25.1); K02734 20S proteasome
subunit beta 4 [EC:3.4.25.1]
Length=199
Score = 37.7 bits (86), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 32/142 (22%), Positives = 62/142 (43%), Gaps = 4/142 (2%)
Query 70 VVCVAGEDFVIGVGDTRLSVGYSIHSRH-HSKITQLTSRCCIASSGMQADINTLHKLLKA 128
++ + G DFV+ D ++ I +H + K+ +L+ + + G D + ++
Sbjct 4 LIGIQGPDFVLVAADN-VAASSIIQMKHDYDKMFKLSEKILLLCVGEAGDTVQFAEYIQK 62
Query 129 RVAVYRHLHRAEPSGPALSQLLSSLL--YSRRFFPFYTFNLLFALDEEGKGAVFGYDAIG 186
V +Y+ + E S A + L Y R P++ LL DE ++ D +
Sbjct 63 NVQLYKMRNGYELSPAAAANFTRKNLADYLRSRTPYHVNLLLAGYDETDGPGLYYMDYLS 122
Query 187 SFERSSFNAAGTGGPLVTALLD 208
+ ++ F A G G L ++LD
Sbjct 123 ALAKAPFAAHGYGAFLTLSILD 144
> xla:443966 psmb2, MGC130720, MGC80364; proteasome (prosome,
macropain) subunit, beta type, 2; K02734 20S proteasome subunit
beta 4 [EC:3.4.25.1]
Length=199
Score = 37.4 bits (85), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 30/141 (21%), Positives = 60/141 (42%), Gaps = 2/141 (1%)
Query 70 VVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKLLKAR 129
++ + G DFV+ DT + + K+ +++ + + G D + ++
Sbjct 4 LIGIQGNDFVLVAADTVCANSIIRMKQDVDKMFKMSEKILLLCVGEAGDTVQFAEYIQKN 63
Query 130 VAVYRHLHRAEPSGPALSQLLSSLL--YSRRFFPFYTFNLLFALDEEGKGAVFGYDAIGS 187
V +Y+ + E S A + L Y R P++ LL DE +++ D + +
Sbjct 64 VQLYKMRNGYELSPTAAANFTRRNLADYLRSRTPYHVNLLLAGYDEHEGPSLYYMDYLAA 123
Query 188 FERSSFNAAGTGGPLVTALLD 208
++ F A G G L ++LD
Sbjct 124 LAKTRFAAHGYGAYLTLSILD 144
> tpv:TP01_0908 proteasome subunit beta type 7 precursor; K02739
20S proteasome subunit beta 2 [EC:3.4.25.1]
Length=262
Score = 37.0 bits (84), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 24/97 (24%), Positives = 47/97 (48%), Gaps = 4/97 (4%)
Query 63 YVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTL 122
Y+ G T+ V G+D V+ DTR + G + ++ SK+ +++ A +G+ AD+
Sbjct 27 YLKTGTTICGVMGKDSVVLAADTRATQGIIVADKNCSKLHKISDNIYCAGAGVAADLEHT 86
Query 123 HKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRF 159
L + ++R + +P +Q+ S+L F
Sbjct 87 TLWLANNIELHRLNTKKQPR----AQMCISMLVHELF 119
> hsa:122706 PSMB11, BETA5T, FLJ16369; proteasome (prosome, macropain)
subunit, beta type, 11 (EC:3.4.25.1); K11598 20S proteasome
subunit beta 11 [EC:3.4.25.1]
Length=300
Score = 37.0 bits (84), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 33/149 (22%), Positives = 59/149 (39%), Gaps = 1/149 (0%)
Query 61 NPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADIN 120
P + +G T + VI DTR S G + K+ + +SG AD
Sbjct 43 GPRLAHGTTTLAFRFRHGVIAAADTRSSCGSYVACPASCKVIPVHQHLLGTTSGTSADCA 102
Query 121 TLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVF 180
T +++L+ + + PS + ++LLS+++ R L D G +
Sbjct 103 TWYRVLQRELRLRELREGQLPSVASAAKLLSAMMSQYRGLDLCVATALCGWDRSGPELFY 162
Query 181 GYDAIGSFERSSFNAAGTGGPLVTALLDN 209
Y + G+ + + G+G P +LD
Sbjct 163 VY-SDGTRLQGDIFSVGSGSPYAYGVLDR 190
> mmu:73902 Psmb11, 5830406J20Rik, beta5t; proteasome (prosome,
macropain) subunit, beta type, 11 (EC:3.4.25.1); K11598 20S
proteasome subunit beta 11 [EC:3.4.25.1]
Length=302
Score = 36.2 bits (82), Expect = 0.084, Method: Compositional matrix adjust.
Identities = 33/149 (22%), Positives = 59/149 (39%), Gaps = 1/149 (0%)
Query 61 NPYVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADIN 120
P + +G T + VI DTR S G + K+ + R +SG AD
Sbjct 43 GPRLAHGTTTLAFRFRHGVIAAADTRSSCGSYVACPASRKVIPVHQRLLGTTSGTSADCA 102
Query 121 TLHKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVF 180
T +++L+ + + PS ++LL++++ R L D G +
Sbjct 103 TWYRVLRRELRLRELREGQLPSVAGTAKLLAAMMSCYRGLDLCVATALCGWDHSGPALFY 162
Query 181 GYDAIGSFERSSFNAAGTGGPLVTALLDN 209
Y + G+ + + G+G P +LD
Sbjct 163 VY-SDGTCLQGDIFSVGSGSPYAYGVLDR 190
> dre:446135 si:ch211-14a17.13 (EC:3.4.25.1); K11598 20S proteasome
subunit beta 11 [EC:3.4.25.1]
Length=375
Score = 35.8 bits (81), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 29/108 (26%), Positives = 50/108 (46%), Gaps = 5/108 (4%)
Query 63 YVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTL 122
++++G T + +D VI DTR S + KI + S + +SG AD
Sbjct 73 HLSHGTTTLGFIFQDGVIAAADTRASCKGLVCCPITHKIMPIHSHLVVNTSGSGADCMLW 132
Query 123 HKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFA 170
++L + +Y+ HR S ++LLS++L+ PF N+ A
Sbjct 133 ERILTREMRLYQLRHRTRLSINGAAKLLSTMLH-----PFKGTNVCVA 175
> dre:100331997 proteasome beta 11 subunit-like
Length=375
Score = 35.8 bits (81), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 29/108 (26%), Positives = 50/108 (46%), Gaps = 5/108 (4%)
Query 63 YVNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTL 122
++++G T + +D VI DTR S + KI + S + +SG AD
Sbjct 73 HLSHGTTTLGFIFQDGVIAAADTRASCKGLVCCPITHKIMPIHSHLVVNTSGSGADCMLW 132
Query 123 HKLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFA 170
++L + +Y+ HR S ++LLS++L+ PF N+ A
Sbjct 133 ERILTREMRLYQLRHRTRLSINGAAKLLSTMLH-----PFKGTNVCVA 175
> dre:100331481 proteasome beta 5 subunit-like
Length=238
Score = 35.4 bits (80), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 34/148 (22%), Positives = 62/148 (41%), Gaps = 1/148 (0%)
Query 64 VNNGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLH 123
+++G T + A + VI DTR S + K+ + S +SG AD
Sbjct 18 LSHGTTTLGFAFQGGVIAAADTRSSCAGKVACPASPKVLPIHSHLVGTTSGTSADCALWK 77
Query 124 KLLKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVFGYD 183
++L + +Y+ HR S ++LLS +L+ + L D G V+
Sbjct 78 RILARELRLYQLRHRRRLSTGGAAKLLSHMLHPFKGTELCVAATLCGWDGGGPRVVY-VC 136
Query 184 AIGSFERSSFNAAGTGGPLVTALLDNQI 211
+ G + + + G+G P ++LD +
Sbjct 137 SDGLRLQGALFSVGSGSPYAYSILDGGV 164
> hsa:5696 PSMB8, D6S216, D6S216E, LMP7, MGC1491, PSMB5i, RING10;
proteasome (prosome, macropain) subunit, beta type, 8 (large
multifunctional peptidase 7) (EC:3.4.25.1); K02740 20S
proteasome subunit beta 8 [EC:3.4.25.1]
Length=272
Score = 35.4 bits (80), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 32/144 (22%), Positives = 61/144 (42%), Gaps = 1/144 (0%)
Query 66 NGGTVVCVAGEDFVIGVGDTRLSVGYSIHSRHHSKITQLTSRCCIASSGMQADINTLHKL 125
+G T + + VI D+R S G I + +K+ ++ SG AD +L
Sbjct 67 HGTTTLAFKFQHGVIAAVDSRASAGSYISALRVNKVIEINPYLLGTMSGCAADCQYWERL 126
Query 126 LKARVAVYRHLHRAEPSGPALSQLLSSLLYSRRFFPFYTFNLLFALDEEGKGAVFGYDAI 185
L +Y + S A S+LLS+++ R +++ D++G G + D
Sbjct 127 LAKECRLYYLRNGERISVSAASKLLSNMMCQYRGMGLSMGSMICGWDKKGPGLYY-VDEH 185
Query 186 GSFERSSFNAAGTGGPLVTALLDN 209
G+ + + G+G ++D+
Sbjct 186 GTRLSGNMFSTGSGNTYAYGVMDS 209
Lambda K H
0.322 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6687805280
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40