bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3211_orf1
Length=209
Score E
Sequences producing significant alignments: (Bits) Value
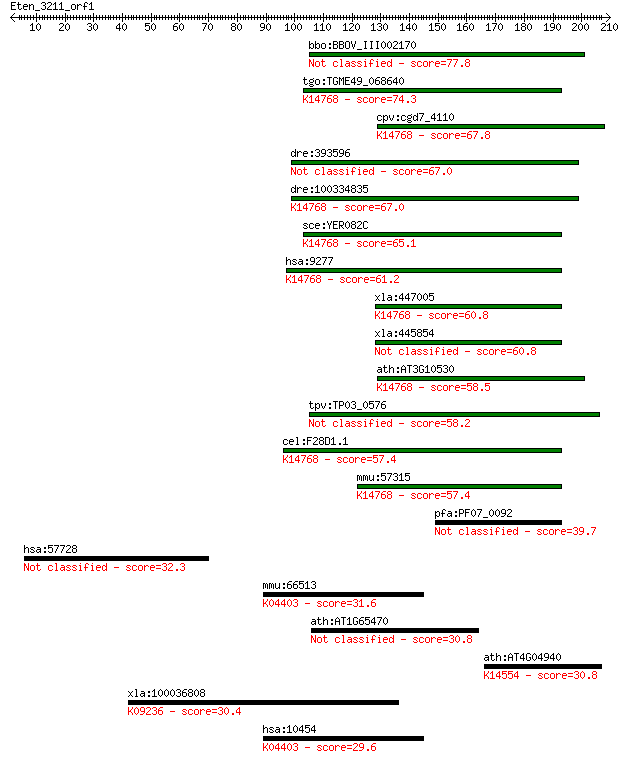
bbo:BBOV_III002170 17.m07209; hypothetical protein 77.8 2e-14
tgo:TGME49_068640 WD domain, G-beta repeat-containing protein ... 74.3 2e-13
cpv:cgd7_4110 WD40 protein (part of U3 processesome) ; K14768 ... 67.8 3e-11
dre:393596 wdr46, Bing4-like, MGC63793, bing4, c6orf11l, zgc:6... 67.0 4e-11
dre:100334835 WD repeat domain 46-like; K14768 U3 small nucleo... 67.0 5e-11
sce:YER082C UTP7, KRE31; Nucleolar protein, component of the s... 65.1 2e-10
hsa:9277 WDR46, BING4, C6orf11, FP221; WD repeat domain 46; K1... 61.2 2e-09
xla:447005 wdr46-a, Bing4, MGC82479, bing4-a, wdr46; WD repeat... 60.8 3e-09
xla:445854 wdr46-b, Bing4, bing4-b; WD repeat domain 46 60.8
ath:AT3G10530 transducin family protein / WD-40 repeat family ... 58.5 1e-08
tpv:TP03_0576 hypothetical protein 58.2 2e-08
cel:F28D1.1 hypothetical protein; K14768 U3 small nucleolar RN... 57.4 3e-08
mmu:57315 Wdr46, 2310007I04Rik, Bing4, C78447, C78559; WD repe... 57.4 4e-08
pfa:PF07_0092 BING4 (NUC141) WD-40 repeat protein, putative 39.7 0.007
hsa:57728 WDR19, DYF-2, FLJ23127, IFT144, KIAA1638, ORF26, Ose... 32.3 1.2
mmu:66513 Tab1, 2310012M03Rik, Map3k7ip1; TGF-beta activated k... 31.6 2.1
ath:AT1G65470 FAS1; FAS1 (FASCIATA 1); histone binding 30.8 3.4
ath:AT4G04940 transducin family protein / WD-40 repeat family ... 30.8 3.5
xla:100036808 tshz3, Xtsh3, teashirt3; teashirt zinc finger ho... 30.4 5.1
hsa:10454 TAB1, 3'-Tab1, MAP3K7IP1, MGC57664; TGF-beta activat... 29.6 8.5
> bbo:BBOV_III002170 17.m07209; hypothetical protein
Length=525
Score = 77.8 bits (190), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 37/96 (38%), Positives = 56/96 (58%), Gaps = 0/96 (0%)
Query 105 SRGTIKRQRALEKKVQESVSAVTALLTETEGYLEAEGLEKTYKFTQADILENVSEEIGKK 164
S+ ++R + L+ + A + L GYLE E E+TY +Q+++L++V +K
Sbjct 28 SKRNVERYKKLKHDALSELEATSVLHINEPGYLEPEEGERTYHISQSELLKSVDVGTKRK 87
Query 165 AFRLGLSFGPYFVDYTRGGRHLLLGGQKREFGCFGL 200
F L LS GPYF +YTR GRH+LLGG K + F +
Sbjct 88 VFNLQLSLGPYFANYTRDGRHMLLGGSKGQLALFDI 123
> tgo:TGME49_068640 WD domain, G-beta repeat-containing protein
; K14768 U3 small nucleolar RNA-associated protein 7
Length=568
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 37/90 (41%), Positives = 54/90 (60%), Gaps = 0/90 (0%)
Query 103 KRSRGTIKRQRALEKKVQESVSAVTALLTETEGYLEAEGLEKTYKFTQADILENVSEEIG 162
K+ +G ++ Q+ L + + +L E EG L EGLEK+Y QADI++
Sbjct 77 KKLKGRLRTQQKLNDEATRRLRTTELMLFEEEGGLLTEGLEKSYAIPQADIVKEADLGTR 136
Query 163 KKAFRLGLSFGPYFVDYTRGGRHLLLGGQK 192
+K F L L FGPY VD++R GRH+L+GG+K
Sbjct 137 EKIFSLDLPFGPYAVDFSRNGRHMLIGGKK 166
> cpv:cgd7_4110 WD40 protein (part of U3 processesome) ; K14768
U3 small nucleolar RNA-associated protein 7
Length=529
Score = 67.8 bits (164), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 34/79 (43%), Positives = 47/79 (59%), Gaps = 0/79 (0%)
Query 129 LLTETEGYLEAEGLEKTYKFTQADILENVSEEIGKKAFRLGLSFGPYFVDYTRGGRHLLL 188
LL ET G+LE EGLEK++ TQ ++ EN+S K F L LS GPY ++Y+R G LL+
Sbjct 58 LLPETPGFLETEGLEKSFSITQNELGENISIGAINKRFSLDLSSGPYSINYSRNGNQLLM 117
Query 189 GGQKREFGCFGLPRNENNL 207
GG+K + N+
Sbjct 118 GGRKGHISMLDISNGAPNI 136
> dre:393596 wdr46, Bing4-like, MGC63793, bing4, c6orf11l, zgc:63793;
WD repeat domain 46
Length=583
Score = 67.0 bits (162), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 39/101 (38%), Positives = 53/101 (52%), Gaps = 1/101 (0%)
Query 99 SRTAKRSRGTIKRQRALEKKVQESVSAVTALLTETEGYLEAEGLEKTYKFTQADILENVS 158
+R + + I +L + Q+ + LL E G+LE + E T Q DI + V
Sbjct 92 ARAGPKLKHAITSSESLSELAQKQAARYGLLLPEDAGFLEGDDGEDTCTIAQDDIADAVD 151
Query 159 EEIGKKAFRLGLS-FGPYFVDYTRGGRHLLLGGQKREFGCF 198
G K F L LS FGPY +DY+R GRHLLLGG++ CF
Sbjct 152 ITSGAKYFNLHLSQFGPYRLDYSRTGRHLLLGGRRGHVACF 192
> dre:100334835 WD repeat domain 46-like; K14768 U3 small nucleolar
RNA-associated protein 7
Length=583
Score = 67.0 bits (162), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 39/101 (38%), Positives = 53/101 (52%), Gaps = 1/101 (0%)
Query 99 SRTAKRSRGTIKRQRALEKKVQESVSAVTALLTETEGYLEAEGLEKTYKFTQADILENVS 158
+R + + I +L + Q+ + LL E G+LE + E T Q DI + V
Sbjct 92 ARAGPKLKHAITSSESLSELAQKQAARYGLLLPEDAGFLEGDDGEDTCTIAQDDIADAVD 151
Query 159 EEIGKKAFRLGLS-FGPYFVDYTRGGRHLLLGGQKREFGCF 198
G K F L LS FGPY +DY+R GRHLLLGG++ CF
Sbjct 152 ITSGAKYFNLHLSQFGPYRLDYSRTGRHLLLGGRRGHVACF 192
> sce:YER082C UTP7, KRE31; Nucleolar protein, component of the
small subunit (SSU) processome containing the U3 snoRNA that
is involved in processing of pre-18S rRNA; K14768 U3 small
nucleolar RNA-associated protein 7
Length=554
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 38/92 (41%), Positives = 51/92 (55%), Gaps = 2/92 (2%)
Query 103 KRSRGTIKRQRALEKKVQESVSAVTALLTETEGYLEAEG-LEKTYKFTQADILENVSEEI 161
K+ R +K+ KK S +A LL E+ GYLE E LEKT+K Q++I +V
Sbjct 40 KKLRAGLKKIDEQYKKAVSSAAATDYLLPESNGYLEPENELEKTFKVQQSEIKSSVDVST 99
Query 162 GKKAFRLGLS-FGPYFVDYTRGGRHLLLGGQK 192
KA L L FGPY + Y + G HLL+ G+K
Sbjct 100 ANKALDLSLKEFGPYHIKYAKNGTHLLITGRK 131
> hsa:9277 WDR46, BING4, C6orf11, FP221; WD repeat domain 46;
K14768 U3 small nucleolar RNA-associated protein 7
Length=556
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 39/97 (40%), Positives = 52/97 (53%), Gaps = 3/97 (3%)
Query 97 PQSRTAKRSRGTIKRQRALEKKVQESVSAVTALLTETEGYLEAEGLEKTYKFTQADILEN 156
P S+ RSR + E ++ + S + LL E G+LE E E T K QADI+E
Sbjct 68 PHSKAKTRSRLEVAEAEEEETSIKAARSEL--LLAEEPGFLEGEDGEDTAKICQADIVEA 125
Query 157 VSEEIGKKAFRLGL-SFGPYFVDYTRGGRHLLLGGQK 192
V K F L L FGPY ++Y+R GRHL GG++
Sbjct 126 VDIASAAKHFDLNLRQFGPYRLNYSRTGRHLAFGGRR 162
> xla:447005 wdr46-a, Bing4, MGC82479, bing4-a, wdr46; WD repeat
domain 46; K14768 U3 small nucleolar RNA-associated protein
7
Length=586
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 33/66 (50%), Positives = 41/66 (62%), Gaps = 1/66 (1%)
Query 128 ALLTETEGYLEAEGLEKTYKFTQADILENVSEEIGKKAFRLGLS-FGPYFVDYTRGGRHL 186
LL E EG+LE + + T TQ DI E V G K F L L+ FGPY ++YTR GR+L
Sbjct 127 VLLPEEEGFLEGDEGQDTCTITQEDIAEIVDITSGSKHFNLNLNQFGPYRINYTRNGRNL 186
Query 187 LLGGQK 192
LL GQ+
Sbjct 187 LLAGQR 192
> xla:445854 wdr46-b, Bing4, bing4-b; WD repeat domain 46
Length=586
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 32/66 (48%), Positives = 40/66 (60%), Gaps = 1/66 (1%)
Query 128 ALLTETEGYLEAEGLEKTYKFTQADILENVSEEIGKKAFRLGLS-FGPYFVDYTRGGRHL 186
LL E EG+LE + + T TQ DI E + G K F L L+ FGPY ++YTR GR L
Sbjct 127 VLLPEEEGFLEGDENQDTCTITQEDIAEVIDITSGSKHFNLNLNQFGPYRINYTRNGRQL 186
Query 187 LLGGQK 192
LL GQ+
Sbjct 187 LLAGQR 192
> ath:AT3G10530 transducin family protein / WD-40 repeat family
protein; K14768 U3 small nucleolar RNA-associated protein
7
Length=536
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 40/73 (54%), Gaps = 1/73 (1%)
Query 129 LLTETEGYLEAEGLEKTYKFTQADILENVSEEIGKKAFRLGL-SFGPYFVDYTRGGRHLL 187
LL GYLE EGLEKT++ Q DI V + + + L FGPY +D+T GRH+L
Sbjct 74 LLPAEAGYLETEGLEKTWRVKQTDIANEVDILSSRNQYDIVLPDFGPYKLDFTASGRHML 133
Query 188 LGGQKREFGCFGL 200
GG+K +
Sbjct 134 AGGRKGHLALLDM 146
> tpv:TP03_0576 hypothetical protein
Length=515
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 36/110 (32%), Positives = 55/110 (50%), Gaps = 10/110 (9%)
Query 105 SRGTIKRQRALEKKVQESVSAVTALLTETEGYLEAEGLEKTYKFTQADILENVSEEIGKK 164
S ++ R + ++ ++ + LL GY+ EKTYK TQ ++L V + KK
Sbjct 26 SNKNFQKNRTICEESIKTTDLTSILLPNEPGYIIPGENEKTYKLTQTNLLTLVDQGTKKK 85
Query 165 AFRLGLSFGPYFVDYTRGGRHLLLGGQKREFGC---------FGLPRNEN 205
L L +GPY VD++ GR+LLLGG+K + F + NEN
Sbjct 86 VI-LNLPYGPYAVDFSHNGRYLLLGGEKGQLSLICTHTYKDFFDISVNEN 134
> cel:F28D1.1 hypothetical protein; K14768 U3 small nucleolar
RNA-associated protein 7
Length=580
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 51/100 (51%), Gaps = 3/100 (3%)
Query 96 DPQSRTAKRSRGTIKRQRALEKKVQESVSAVTALLTETEGYL--EAEGLEKTYKFTQADI 153
DP S +K + +K + ++ E+ + L E G++ + E TY Q DI
Sbjct 90 DPSSMRSKLQQEKMKVTKDKFQRRIEATAKAEKRLVENTGFVAPDEEDHSLTYSIRQKDI 149
Query 154 LENVSEEIGKKAFRLGLS-FGPYFVDYTRGGRHLLLGGQK 192
E+V K F L L FGPY +DYT GRHL++GG+K
Sbjct 150 AESVDLAAATKHFELKLPRFGPYHIDYTDNGRHLVIGGRK 189
> mmu:57315 Wdr46, 2310007I04Rik, Bing4, C78447, C78559; WD repeat
domain 46; K14768 U3 small nucleolar RNA-associated protein
7
Length=622
Score = 57.4 bits (137), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 35/74 (47%), Positives = 42/74 (56%), Gaps = 3/74 (4%)
Query 122 SVSAVTA--LLTETEGYLEAEGLEKTYKFTQADILENVSEEIGKKAFRLGL-SFGPYFVD 178
SV A A LL E G+L E E T K Q DI+E V K F L L FGPY ++
Sbjct 142 SVRAARAELLLAEEPGFLVGEDGEDTAKILQTDIVEAVDIASAAKHFDLNLRQFGPYRLN 201
Query 179 YTRGGRHLLLGGQK 192
Y+R GRHL LGG++
Sbjct 202 YSRTGRHLALGGRR 215
> pfa:PF07_0092 BING4 (NUC141) WD-40 repeat protein, putative
Length=602
Score = 39.7 bits (91), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 149 TQADILENVSEEIGKKAFRLGLSFGPYFVDYTRGGRHLLLGGQK 192
+Q I +N KK F L L+ GPY YTR G++LL+ G K
Sbjct 201 SQKYIYDNADVGTQKKVFDLKLNMGPYSCTYTRNGKYLLMTGVK 244
> hsa:57728 WDR19, DYF-2, FLJ23127, IFT144, KIAA1638, ORF26, Oseg6,
PWDMP; WD repeat domain 19
Length=1342
Score = 32.3 bits (72), Expect = 1.2, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 25/64 (39%), Gaps = 10/64 (15%)
Query 6 HLFLSSWQAPDTDSLRPQRAPGCLRGSFCDLGVHCLAQNEAATWLALAVGLQQPNQLQRG 65
H FLS+ + D LRP A + F D C N+ A W N+L R
Sbjct 635 HGFLSNLKDTGPDELRPMLAQNLMLKRFSDAWEMCRILNDEAAW----------NELARA 684
Query 66 ALHQ 69
LH
Sbjct 685 CLHH 688
> mmu:66513 Tab1, 2310012M03Rik, Map3k7ip1; TGF-beta activated
kinase 1/MAP3K7 binding protein 1; K04403 TAK1-binding protein
1
Length=502
Score = 31.6 bits (70), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 31/56 (55%), Gaps = 0/56 (0%)
Query 89 QSQLPMDDPQSRTAKRSRGTIKRQRALEKKVQESVSAVTALLTETEGYLEAEGLEK 144
QSQLP PQ + + + ++R +ALE+++ AV A+L ++ Y+ G +
Sbjct 132 QSQLPEGVPQHQLPPQYQKILERLKALEREISGGAMAVVAVLLNSKLYVANVGTNR 187
> ath:AT1G65470 FAS1; FAS1 (FASCIATA 1); histone binding
Length=815
Score = 30.8 bits (68), Expect = 3.4, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 33/58 (56%), Gaps = 8/58 (13%)
Query 106 RGTIKRQRALEKKVQESVSAVTALLTETEGYLEAEGLEKTYKFTQADILENVSEEIGK 163
RG +K +R KK+ E ++AV+A+L + E+T K ++D L +E++GK
Sbjct 165 RGVLKLRRTCRKKIHERITAVSAMLAALQ-------REETEKLWRSD-LSKAAEKLGK 214
> ath:AT4G04940 transducin family protein / WD-40 repeat family
protein; K14554 U3 small nucleolar RNA-associated protein
21
Length=910
Score = 30.8 bits (68), Expect = 3.5, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 166 FRLGLSFGPYFVDYTRGGRHLLLGGQKREFGCFGLPRNENN 206
FR G S P + + GRH+L GQ R F F + + + +
Sbjct 309 FRSGHSAPPLCIRFYSNGRHILSAGQDRAFRLFSVIQEQQS 349
> xla:100036808 tshz3, Xtsh3, teashirt3; teashirt zinc finger
homeobox 3; K09236 teashirt
Length=1004
Score = 30.4 bits (67), Expect = 5.1, Method: Composition-based stats.
Identities = 27/99 (27%), Positives = 43/99 (43%), Gaps = 7/99 (7%)
Query 42 AQNEAATW---LALAVGLQQPN--QLQRGALHQARDMGGPQGAQASTAPGAKQSQLPMDD 96
AQN +W ++ Q PN +L G+ +A + G+ +P ++Q M
Sbjct 462 AQNGTPSWGGYPSIHAAYQLPNMMKLSLGSSGKATPLKPMYGSSELISP--TKNQPIMSP 519
Query 97 PQSRTAKRSRGTIKRQRALEKKVQESVSAVTALLTETEG 135
P S+T+ + L KKV E V+ V + E EG
Sbjct 520 PSSQTSPVPKTNFHAMEELVKKVTEKVAKVEEKMKEPEG 558
> hsa:10454 TAB1, 3'-Tab1, MAP3K7IP1, MGC57664; TGF-beta activated
kinase 1/MAP3K7 binding protein 1; K04403 TAK1-binding
protein 1
Length=504
Score = 29.6 bits (65), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 29/56 (51%), Gaps = 0/56 (0%)
Query 89 QSQLPMDDPQSRTAKRSRGTIKRQRALEKKVQESVSAVTALLTETEGYLEAEGLEK 144
QSQLP PQ + + + ++R + LE+++ AV A+L + Y+ G +
Sbjct 132 QSQLPEGVPQHQLPPQYQKILERLKTLEREISGGAMAVVAVLLNNKLYVANVGTNR 187
Lambda K H
0.316 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6494887820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40