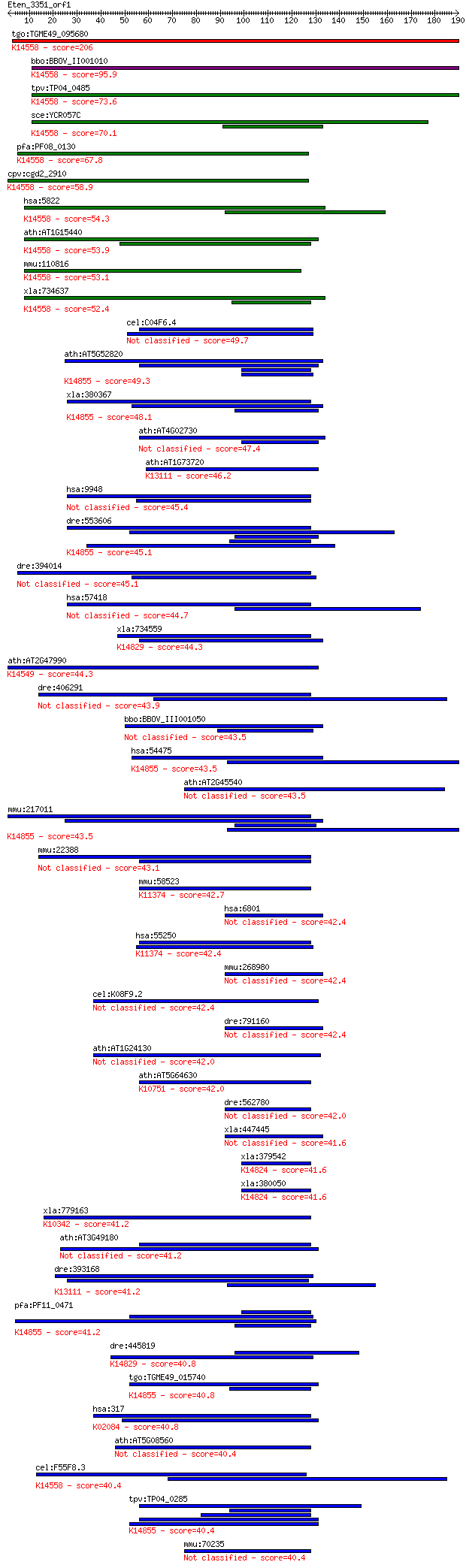
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3351_orf1
Length=189
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_095680 periodic tryptophan protein PWP2, putative ;... 206 3e-53
bbo:BBOV_II001010 18.m06073; periodic tryptophan protein 2-lik... 95.9 8e-20
tpv:TP04_0485 hypothetical protein; K14558 periodic tryptophan... 73.6 4e-13
sce:YCR057C PWP2, UTP1, YCR055C, YCR058C; Conserved 90S pre-ri... 70.1 4e-12
pfa:PF08_0130 WD-repeat protein, putative; K14558 periodic try... 67.8 2e-11
cpv:cgd2_2910 hypothetical protein ; K14558 periodic tryptopha... 58.9 9e-09
hsa:5822 PWP2, EHOC-17, PWP2H; PWP2 periodic tryptophan protei... 54.3 3e-07
ath:AT1G15440 transducin family protein / WD-40 repeat family ... 53.9 3e-07
mmu:110816 Pwp2, 6530411D08Rik, MGC117973, Pwp2h, wdp103; PWP2... 53.1 5e-07
xla:734637 hypothetical protein MGC115367; K14558 periodic try... 52.4 9e-07
cel:C04F6.4 unc-78; UNCoordinated family member (unc-78) 49.7 7e-06
ath:AT5G52820 WD-40 repeat family protein / notchless protein,... 49.3 8e-06
xla:380367 nle1, MGC52549, nle; notchless homolog 1; K14855 ri... 48.1 2e-05
ath:AT4G02730 transducin family protein / WD-40 repeat family ... 47.4 3e-05
ath:AT1G73720 transducin family protein / WD-40 repeat family ... 46.2 7e-05
hsa:9948 WDR1, AIP1, NORI-1; WD repeat domain 1 45.4
dre:553606 nle1, MGC110281, fc12b09, fc45f01, wu:fc12b09, wu:f... 45.1 1e-04
dre:394014 wdr1, MGC55793, wu:fa66e09, zgc:55793, zgc:77547; W... 45.1 2e-04
hsa:57418 WDR18, MGC2436, R32184_1; WD repeat domain 18 44.7
xla:734559 wdr18, MGC115442; WD repeat domain 18; K14829 pre-r... 44.3 2e-04
ath:AT2G47990 SWA1; SWA1 (SLOW WALKER1); nucleotide binding; K... 44.3 2e-04
dre:406291 katnb1, wu:fj32f02, wu:fj65h01, zgc:56071; katanin ... 43.9 4e-04
bbo:BBOV_III001050 17.m07120; WD domain, G-beta repeat contain... 43.5 4e-04
hsa:54475 NLE1, FLJ10458, Nle; notchless homolog 1 (Drosophila... 43.5 4e-04
ath:AT2G45540 WD-40 repeat family protein / beige-related 43.5 4e-04
mmu:217011 Nle1, AL022765, BC018399, MGC25690, Nle; notchless ... 43.5 5e-04
mmu:22388 Wdr1, Aip1, D5Wsu185e, rede; WD repeat domain 1 43.1
mmu:58523 Elp2, AU023723, Epl2, StIP1, Statip1; elongation pro... 42.7 8e-04
hsa:6801 STRN, MGC125642, SG2NA; striatin, calmodulin binding ... 42.4 8e-04
hsa:55250 ELP2, FLJ10879, SHINC-2, STATIP1, StIP; elongation p... 42.4 9e-04
mmu:268980 Strn, AU022939, D130055P19; striatin, calmodulin bi... 42.4 9e-04
cel:K08F9.2 tag-216; Temporarily Assigned Gene name family mem... 42.4 0.001
dre:791160 strn; zgc:158357 42.4 0.001
ath:AT1G24130 transducin family protein / WD-40 repeat family ... 42.0 0.001
ath:AT5G64630 FAS2; FAS2 (FASCIATA 2); nucleotide binding / pr... 42.0 0.001
dre:562780 strn4; striatin, calmodulin binding protein 4 42.0
xla:447445 strn, MGC81438; striatin, calmodulin binding protein 41.6 0.002
xla:379542 bop1-a, MGC68939; ribosome biogenesis protein bop1-... 41.6 0.002
xla:380050 bop1, MGC52658, bop1-B; block of proliferation 1; K... 41.6 0.002
xla:779163 wsb2, MGC83316; WD repeat and SOCS box containing 2... 41.2 0.002
ath:AT3G49180 RID3; RID3 (ROOT INITIATION DEFECTIVE 3); nucleo... 41.2 0.002
dre:393168 smu1, MGC56147, zgc:56147; smu-1 suppressor of mec-... 41.2 0.002
pfa:PF11_0471 nucleolar preribosomal assembly protein, putativ... 41.2 0.002
dre:445819 wdr18, wu:fa16f06, wu:fb79d07, zgc:92370; WD repeat... 40.8 0.002
tgo:TGME49_015740 hypothetical protein ; K14855 ribosome assem... 40.8 0.003
hsa:317 APAF1, APAF-1, CED4, DKFZp781B1145; apoptotic peptidas... 40.8 0.003
ath:AT5G08560 transducin family protein / WD-40 repeat family ... 40.4 0.003
cel:F55F8.3 hypothetical protein; K14558 periodic tryptophan p... 40.4 0.004
tpv:TP04_0285 hypothetical protein; K14855 ribosome assembly p... 40.4 0.004
mmu:70235 Poc1a, 2510040D07Rik, Wdr51a; POC1 centriolar protei... 40.4 0.004
> tgo:TGME49_095680 periodic tryptophan protein PWP2, putative
; K14558 periodic tryptophan protein 2
Length=1266
Score = 206 bits (525), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 111/221 (50%), Positives = 135/221 (61%), Gaps = 34/221 (15%)
Query 3 FCIRSDSQLAISIDVHGQGLVINLAKGNVLNRIRFKSNTTS--KIRWQQQKQ-QHLVTAA 59
F +RSD+ LAI ID+HGQG+V+NLAKG V+NRI+FKSNT++ K +W K QH VT A
Sbjct 116 FAVRSDNLLAICIDLHGQGMVVNLAKGCVVNRIQFKSNTSAEQKRKWDAVKDLQHSVTGA 175
Query 60 AFSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGS 119
AFSPDD FAVA GRS+Q+WH+P+A YQL LL S TLHQQ IT LSWSPDS HLV+GS
Sbjct 176 AFSPDDRFFAVAAGRSVQIWHAPTARHAYQLRLLRSMTLHQQTITTLSWSPDSTHLVTGS 235
Query 120 QDCTVRLWRAR-------------------------------GKWGGQTSAAAAAAAATA 148
DCTVRLWRA+ GG A +
Sbjct 236 LDCTVRLWRAKVAKSGLVSSSVSGNGGSLEGLVVRDEDESAARPDGGVLEAETGYSFPRD 295
Query 149 ATAGTEEAAAAAGDAAFIPSAFLSHRHELKFVCFSPCLTRI 189
+ ++ A GD FIP AF+SH++ L+F CFS CLTRI
Sbjct 296 SGRKADKGDATCGDNEFIPVAFVSHKYPLRFACFSGCLTRI 336
> bbo:BBOV_II001010 18.m06073; periodic tryptophan protein 2-like
protein; K14558 periodic tryptophan protein 2
Length=988
Score = 95.9 bits (237), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 60/185 (32%), Positives = 86/185 (46%), Gaps = 36/185 (19%)
Query 11 LAISIDVHGQGLVINLAKGNVLNRIRFKSNTT------SKIRWQQQKQQHLVTAAAFSPD 64
L I IDVHG G ++NL + +L+R++FK+++T S + Q LV AAFSPD
Sbjct 68 LVILIDVHGYGYILNLLRDCILHRLQFKTSSTVINEKRSTSVLPANESQRLVRDAAFSPD 127
Query 65 DALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTV 124
FAVA G + +W +P ++++ L T H I+ + WS DS + + S D TV
Sbjct 128 ARFFAVAIGNKLCIWRAPEEGLSWRMTLHRELTGHMDTISSVDWSSDSRFICTASADLTV 187
Query 125 RLWRARGKWGGQTSAAAAAAAATAATAGTEEAAAAAGDAAFIPSAFLSHRHELKFVCFSP 184
RLW G F+P AF+ HR +K FS
Sbjct 188 RLWSVDPIPG------------------------------FVPCAFVDHRRPVKTAFFSS 217
Query 185 CLTRI 189
+TRI
Sbjct 218 DMTRI 222
> tpv:TP04_0485 hypothetical protein; K14558 periodic tryptophan
protein 2
Length=968
Score = 73.6 bits (179), Expect = 4e-13, Method: Composition-based stats.
Identities = 50/194 (25%), Positives = 83/194 (42%), Gaps = 45/194 (23%)
Query 11 LAISIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQQ---------------QKQQHL 55
L+I +D +G G ++NL + +++R++FKS+T + Q+
Sbjct 68 LSILVDDNGFGYILNLIRDKIVHRLQFKSSTNISTAESKGYNSLKLVFSYIIILSTQKKR 127
Query 56 VTAAAFSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHL 115
+ A+FSP FAV+ + + +W SP ++++ L T H I L WS DS ++
Sbjct 128 LIHASFSPYGKYFAVSIQKKLLVWLSPDEKLSWRMTLHLEITGHMDNINSLDWSKDSKYI 187
Query 116 VSGSQDCTVRLWRARGKWGGQTSAAAAAAAATAATAGTEEAAAAAGDAAFIPSAFLSHRH 175
+ S+D TVRLW D F+P AF+ HR
Sbjct 188 CTSSKDLTVRLW------------------------------CVEPDDTFVPVAFVDHRR 217
Query 176 ELKFVCFSPCLTRI 189
+K F+ + RI
Sbjct 218 SVKAAFFTDDMNRI 231
> sce:YCR057C PWP2, UTP1, YCR055C, YCR058C; Conserved 90S pre-ribosomal
component essential for proper endonucleolytic cleavage
of the 35 S rRNA precursor at A0, A1, and A2 sites; contains
eight WD-repeats; PWP2 deletion leads to defects in cell
cycle and bud morphogenesis; K14558 periodic tryptophan
protein 2
Length=923
Score = 70.1 bits (170), Expect = 4e-12, Method: Composition-based stats.
Identities = 49/173 (28%), Positives = 73/173 (42%), Gaps = 44/173 (25%)
Query 11 LAISIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSPDDALFAV 70
L ISID G+ +++N NVL+ FK +A FSPD LFA+
Sbjct 69 LLISIDEDGRAILVNFKARNVLHHFNFKEK---------------CSAVKFSPDGRLFAL 113
Query 71 ACGRSIQLWHSPSAATNYQLHLLTSFTLHQ---QLITCLSWSPDSLHLVSGSQDCTVRLW 127
A GR +Q+W +P + Q +H Q IT L+WS DS +++ S+D + ++W
Sbjct 114 ASGRFLQIWKTPDVNKDRQFAPFVRHRVHAGHFQDITSLTWSQDSRFILTTSKDLSAKIW 173
Query 128 RARGKWGGQTSAAAAAAAATAATAGTEEAAAAA----GDAAFIPSAFLSHRHE 176
+ +EE AA G ++ AF SH E
Sbjct 174 ----------------------SVDSEEKNLAATTFNGHRDYVMGAFFSHDQE 204
Score = 34.3 bits (77), Expect = 0.28, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 26/42 (61%), Gaps = 0/42 (0%)
Query 91 HLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRARGK 132
LL + + H+ ++CLS+S ++ L S S D T+R+W G+
Sbjct 464 QLLDALSGHEGPVSCLSFSQENSVLASASWDKTIRIWSIFGR 505
> pfa:PF08_0130 WD-repeat protein, putative; K14558 periodic tryptophan
protein 2
Length=1121
Score = 67.8 bits (164), Expect = 2e-11, Method: Composition-based stats.
Identities = 38/128 (29%), Positives = 61/128 (47%), Gaps = 6/128 (4%)
Query 5 IRSDSQLAISIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQ------QQKQQHLVTA 58
I ++ I+ID G G VINL K +++RI FKS T + Q++Q +V +
Sbjct 62 IHPSMEIIITIDKFGYGCVINLLKDQIISRILFKSKTGVVTSFNYNNMLTPQEEQDVVNS 121
Query 59 AAFSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSG 118
A F+ F + GR + +W SP NY+L H + + S D + ++
Sbjct 122 AMFTNSGKYFLITVGRRVVIWKSPCKQNNYKLIKYNDICFHSLNVISIDISTDDKYFLTT 181
Query 119 SQDCTVRL 126
S D T+R+
Sbjct 182 SYDMTIRI 189
> cpv:cgd2_2910 hypothetical protein ; K14558 periodic tryptophan
protein 2
Length=1003
Score = 58.9 bits (141), Expect = 9e-09, Method: Composition-based stats.
Identities = 35/126 (27%), Positives = 55/126 (43%), Gaps = 15/126 (11%)
Query 1 STFCIRSDSQLAISIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAA 60
S C+ D + I +D G ++N +KG +LNR++F + V
Sbjct 56 SKMCLSPDESILICVDEDNYGSIVNFSKGLILNRMQFPGS---------------VGFIC 100
Query 61 FSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQ 120
+S + A A I +WH+P + +QL L +H + L WS L++ S
Sbjct 101 YSHNGNYVAAAIDSGIYIWHAPCISKGWQLILKRKHIIHNSKVNSLDWSRCDRFLLTASN 160
Query 121 DCTVRL 126
D TVRL
Sbjct 161 DMTVRL 166
> hsa:5822 PWP2, EHOC-17, PWP2H; PWP2 periodic tryptophan protein
homolog (yeast); K14558 periodic tryptophan protein 2
Length=919
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 37/128 (28%), Positives = 61/128 (47%), Gaps = 19/128 (14%)
Query 8 DSQLAISIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSPDDAL 67
D +LAI +D G L+++L +VL+ FK + V + +FSPD
Sbjct 66 DGRLAIIVDEGGDALLVSLVCRSVLHHFHFKGS---------------VHSVSFSPDGRK 110
Query 68 FAVACGRSIQLWHSPSAATNYQLHLL--TSFTLHQQLITCLSWSPDSLHLVSGSQDCTVR 125
F V G Q++H+P + +L T F + + TC+ W+ DS V GS+D +
Sbjct 111 FVVTKGNIAQMYHAPGKKREFNAFVLDKTYFGPYDE-TTCIDWTDDSRCFVVGSKDMSTW 169
Query 126 LWRARGKW 133
++ A +W
Sbjct 170 VFGAE-RW 176
Score = 30.0 bits (66), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 32/69 (46%), Gaps = 2/69 (2%)
Query 92 LLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRARGKWGGQTSAA--AAAAAATAA 149
LL + H+ I+ L ++P L S S D TVRLW W + + A + A A T
Sbjct 493 LLDVLSGHEGPISGLCFNPMKSVLASASWDKTVRLWDMFDSWRTKETLALTSDALAVTFR 552
Query 150 TAGTEEAAA 158
G E A A
Sbjct 553 PDGAELAVA 561
> ath:AT1G15440 transducin family protein / WD-40 repeat family
protein; K14558 periodic tryptophan protein 2
Length=900
Score = 53.9 bits (128), Expect = 3e-07, Method: Composition-based stats.
Identities = 36/124 (29%), Positives = 54/124 (43%), Gaps = 16/124 (12%)
Query 8 DSQLAISIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSPDDAL 67
D +++D + L INL + VL+RI FK V A FSP+
Sbjct 63 DGTFLLAVDEQNRCLFINLPRRVVLHRITFKDK---------------VGALKFSPNGKF 107
Query 68 FAVACGRSIQLWHSPS-AATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRL 126
AV G+ +++W SP + +F + L WS DS +L+ GS+D RL
Sbjct 108 IAVGIGKLVEIWRSPGFRRAVLPFERVRTFANSDDKVVSLEWSLDSDYLLVGSRDLAARL 167
Query 127 WRAR 130
+ R
Sbjct 168 FCVR 171
Score = 38.9 bits (89), Expect = 0.011, Method: Composition-based stats.
Identities = 25/81 (30%), Positives = 36/81 (44%), Gaps = 7/81 (8%)
Query 48 QQQKQQHLVTAAAFSPDDALFAV-ACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCL 106
+QQ V +SPD L A A +++W+ S +FT H +T L
Sbjct 384 KQQGHYFDVNCVTYSPDSQLLATGADDNKVKVWNVMSGTC------FITFTEHTNAVTAL 437
Query 107 SWSPDSLHLVSGSQDCTVRLW 127
+ D+ L+S S D TVR W
Sbjct 438 HFMADNHSLLSASLDGTVRAW 458
> mmu:110816 Pwp2, 6530411D08Rik, MGC117973, Pwp2h, wdp103; PWP2
periodic tryptophan protein homolog (yeast); K14558 periodic
tryptophan protein 2
Length=919
Score = 53.1 bits (126), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 56/118 (47%), Gaps = 18/118 (15%)
Query 8 DSQLAISIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSPDDAL 67
D +LAI +D G L+++L +VL+ FK + V + +FSPD
Sbjct 66 DGRLAIIVDEGGAALLVSLVCRSVLHHFHFKGS---------------VHSVSFSPDGRK 110
Query 68 FAVACGRSIQLWHSPSAATNYQLHLL--TSFTLHQQLITCLSWSPDSLHLVSGSQDCT 123
F V G Q++H+P + +L T F + + TC+ W+ DS V GS+D +
Sbjct 111 FVVTKGNIAQMYHAPGKKREFNAFVLDKTYFGPYDE-TTCIDWTDDSKCFVVGSKDMS 167
> xla:734637 hypothetical protein MGC115367; K14558 periodic tryptophan
protein 2
Length=895
Score = 52.4 bits (124), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 35/127 (27%), Positives = 55/127 (43%), Gaps = 17/127 (13%)
Query 8 DSQLAISIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSPDDAL 67
D L I +D G L+I+LA ++LN+ F Q V + FSPD
Sbjct 66 DGSLGILVDEEGAALLISLATKSLLNQFSF---------------QQPVQSVRFSPDGKK 110
Query 68 FAVACGRSIQLWHSPSAATNYQLHLL-TSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRL 126
F + G L+H+P + +L +F TC+ W+ DS GS+D + +
Sbjct 111 FVITKGNVAMLYHAPGKRREFNAFVLDKTFYGPYDETTCIDWTDDSKCFAVGSKDMSTWV 170
Query 127 WRARGKW 133
+ A +W
Sbjct 171 FGAE-RW 176
Score = 32.3 bits (72), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 22/37 (59%), Gaps = 4/37 (10%)
Query 95 SFTLHQQ----LITCLSWSPDSLHLVSGSQDCTVRLW 127
S+ L QQ + LS+SPD H+V+G D V++W
Sbjct 356 SYVLKQQGHFNNMGSLSYSPDGQHIVTGGDDGKVKVW 392
> cel:C04F6.4 unc-78; UNCoordinated family member (unc-78)
Length=611
Score = 49.7 bits (117), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 44/73 (60%), Gaps = 2/73 (2%)
Query 56 VTAAAFSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHL 115
+T+ AFS ++ F VA +S ++ S A N++L S+T H + C+SWSPD++ L
Sbjct 494 ITSVAFS-NNGAFLVATDQSRKVIPY-SVANNFELAHTNSWTFHTAKVACVSWSPDNVRL 551
Query 116 VSGSQDCTVRLWR 128
+GS D +V +W
Sbjct 552 ATGSLDNSVIVWN 564
Score = 36.2 bits (82), Expect = 0.064, Method: Compositional matrix adjust.
Identities = 20/80 (25%), Positives = 35/80 (43%), Gaps = 2/80 (2%)
Query 51 KQQHLVTAAAFSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSF--TLHQQLITCLSW 108
+ V + ++PD +LFA G + ++ T + S H + L+W
Sbjct 188 EHTKFVHSVRYNPDGSLFASTGGDGTIVLYNGVDGTKTGVFEDDSLKNVAHSGSVFGLTW 247
Query 109 SPDSLHLVSGSQDCTVRLWR 128
SPD + S S D T+++W
Sbjct 248 SPDGTKIASASADKTIKIWN 267
> ath:AT5G52820 WD-40 repeat family protein / notchless protein,
putative; K14855 ribosome assembly protein 4
Length=473
Score = 49.3 bits (116), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 37/119 (31%), Positives = 56/119 (47%), Gaps = 17/119 (14%)
Query 25 NLAKGNVLNRIRFKSNTTSKIRWQ-----QQKQ-----QHLVTAAAFSPDDALFAVAC-G 73
N KG+ R+ S+ + W+ Q K+ Q LV FSPD A A
Sbjct 322 NKTKGDSPERLVSGSDDFTMFLWEPSVSKQPKKRLTGHQQLVNHVYFSPDGKWIASASFD 381
Query 74 RSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRARGK 132
+S++LW+ + +T F H + +SWS DS L+SGS+D T+++W R K
Sbjct 382 KSVRLWNGITG------QFVTVFRGHVGPVYQVSWSADSRLLLSGSKDSTLKIWEIRTK 434
Score = 37.4 bits (85), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 16/29 (55%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 99 HQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
H + + C+S+SPD L SGS D TVRLW
Sbjct 108 HAEAVLCVSFSPDGKQLASGSGDTTVRLW 136
Score = 37.4 bits (85), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 35/76 (46%), Gaps = 7/76 (9%)
Query 56 VTAAAFSPDDALFAVACGRS-IQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLH 114
V +FSPD A G + ++LW Y L + H+ + ++WSPD H
Sbjct 112 VLCVSFSPDGKQLASGSGDTTVRLWDL------YTETPLFTCKGHKNWVLTVAWSPDGKH 165
Query 115 LVSGSQDCTVRLWRAR 130
LVSGS+ + W +
Sbjct 166 LVSGSKSGEICCWNPK 181
Score = 34.7 bits (78), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 20/30 (66%), Gaps = 1/30 (3%)
Query 99 HQQLITCLSWSPDSLHLVSGSQDCTVRLWR 128
H +TC+ W D + + +GSQDCT+++W
Sbjct 240 HTLAVTCVKWGGDGI-IYTGSQDCTIKMWE 268
> xla:380367 nle1, MGC52549, nle; notchless homolog 1; K14855
ribosome assembly protein 4
Length=476
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 51/108 (47%), Gaps = 11/108 (10%)
Query 26 LAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSPDDALFAVACGRS-IQLWHSPSA 84
LA G+ +RF +T + + H V + A+SPD A C S I +W PS
Sbjct 120 LASGSGDTTVRFWDLSTETPHFTSKGHTHWVLSIAWSPDGKKLASGCKNSQIFIW-DPST 178
Query 85 ATNYQLHLLTSFTLHQQLITCLSW-----SPDSLHLVSGSQDCTVRLW 127
+ T H + IT L W +P+S +L S S+DCT+R+W
Sbjct 179 GK----QIGKPLTGHSKWITWLCWEPLHLNPESRYLASASKDCTIRIW 222
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 42/81 (51%), Gaps = 7/81 (8%)
Query 53 QHLVTAAAFSPDDALFAVAC-GRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPD 111
Q L+ FSPD + A A +SI+LW + LTS H + ++WS D
Sbjct 363 QALINEVLFSPDTRIIASASFDKSIKLWDGKTG------KFLTSLRGHVSAVYQIAWSAD 416
Query 112 SLHLVSGSQDCTVRLWRARGK 132
S LVSGS D T+++W ++ K
Sbjct 417 SRLLVSGSSDSTLKVWDSKTK 437
Score = 31.6 bits (70), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 21/35 (60%), Gaps = 1/35 (2%)
Query 96 FTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRAR 130
T H Q +T + W D L L S SQD T++ WRA+
Sbjct 233 LTSHTQSVTAVKWGGDGL-LYSSSQDRTIKAWRAQ 266
> ath:AT4G02730 transducin family protein / WD-40 repeat family
protein
Length=333
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 43/79 (54%), Gaps = 7/79 (8%)
Query 56 VTAAAFSPDDALFAVA-CGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLH 114
++ FS D L A A +++ LW +ATNY L + + H I+ L+WS DS +
Sbjct 46 ISCVKFSNDGNLLASASVDKTMILW----SATNYSL--IHRYEGHSSGISDLAWSSDSHY 99
Query 115 LVSGSQDCTVRLWRARGKW 133
S S DCT+R+W AR +
Sbjct 100 TCSASDDCTLRIWDARSPY 118
Score = 33.1 bits (74), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 99 HQQLITCLSWSPDSLHLVSGSQDCTVRLWRAR 130
H + C++++P S +VSGS D T+R+W +
Sbjct 127 HTNFVFCVNFNPPSNLIVSGSFDETIRIWEVK 158
> ath:AT1G73720 transducin family protein / WD-40 repeat family
protein; K13111 WD40 repeat-containing protein SMU1
Length=511
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 37/75 (49%), Gaps = 3/75 (4%)
Query 59 AAFSPDDALFAVACGRS-IQLWH--SPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHL 115
A FSPD A + I++W S + Q SF +H + C+ +S DS L
Sbjct 219 ARFSPDGQFLASSSVDGFIEVWDYISGKLKKDLQYQADESFMMHDDPVLCIDFSRDSEML 278
Query 116 VSGSQDCTVRLWRAR 130
SGSQD +++WR R
Sbjct 279 ASGSQDGKIKIWRIR 293
> hsa:9948 WDR1, AIP1, NORI-1; WD repeat domain 1
Length=466
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 36/104 (34%), Positives = 49/104 (47%), Gaps = 6/104 (5%)
Query 26 LAKGNVLNRIRFKS--NTTSKIRWQQQKQQHLVTAAAFSPDDALFAVACGRSIQLWHSPS 83
+A G V +R S TT K + + + VT A+S D A AV C S ++ S
Sbjct 320 VAIGGVDGNVRLYSILGTTLKDEGKLLEAKGPVTDVAYSHDGAFLAV-CDAS-KVVTVFS 377
Query 84 AATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
A Y + + F H I CL+WSPD+ H SG D V +W
Sbjct 378 VADGYSENNV--FYGHHAKIVCLAWSPDNEHFASGGMDMMVYVW 419
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 34/74 (45%), Gaps = 4/74 (5%)
Query 55 LVTAAAFSPDDALFAVACGRS-IQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSL 113
V FSPD FA A I ++ T ++ L H I +SWSPDS
Sbjct 51 FVNCVRFSPDGNRFATASADGQIYIY---DGKTGEKVCALGGSKAHDGGIYAISWSPDST 107
Query 114 HLVSGSQDCTVRLW 127
HL+S S D T ++W
Sbjct 108 HLLSASGDKTSKIW 121
> dre:553606 nle1, MGC110281, fc12b09, fc45f01, wu:fc12b09, wu:fc45f01,
zgc:110281; notchless homolog 1 (Drosophila); K14855
ribosome assembly protein 4
Length=476
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 36/108 (33%), Positives = 49/108 (45%), Gaps = 11/108 (10%)
Query 26 LAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSPDDALFAVACGRS-IQLWHSPSA 84
LA G+ +RF +T + H V + A+SPD A C S I LW
Sbjct 120 LASGSGDTTVRFWDLSTETPHHTSRGHTHWVLSIAWSPDGKKLASGCKNSQIFLW---DP 176
Query 85 ATNYQLHLLTSFTLHQQLITCLSWSPDSL-----HLVSGSQDCTVRLW 127
T Q+ + T H + IT L W P L +L S S+DCT+R+W
Sbjct 177 VTGKQIG--KTLTGHTKWITWLCWEPLHLNPECRYLASTSKDCTIRIW 222
Score = 43.5 bits (101), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 34/113 (30%), Positives = 48/113 (42%), Gaps = 8/113 (7%)
Query 52 QQHLVTAAAFSPDDALFAVAC-GRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSP 110
Q LV FSPD L A A +SI++W + L S H + ++WS
Sbjct 362 HQALVNEVLFSPDTRLIASASFDKSIKIWDGKTGK------YLNSLRGHVGPVYQVAWSA 415
Query 111 DSLHLVSGSQDCTVRLWRAR-GKWGGQTSAAAAAAAATAATAGTEEAAAAAGD 162
DS LVSGS D T+++W + GK A A + + A+ D
Sbjct 416 DSRLLVSGSSDSTLKVWDIKTGKLNADLPGHADEVFAVDWSPDGQRVASGGKD 468
Score = 34.3 bits (77), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Query 96 FTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRAR 130
T H +TC+ W D L L + SQD T+++WRA+
Sbjct 233 LTGHTHSVTCVKWGGDGL-LYTSSQDRTIKVWRAK 266
Score = 29.6 bits (65), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 94 TSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
+S H + + +++SP +L SGS D TVR W
Sbjct 99 SSLEGHTEAVISVAFSPTGKYLASGSGDTTVRFW 132
Score = 29.3 bits (64), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 26/106 (24%), Positives = 45/106 (42%), Gaps = 12/106 (11%)
Query 34 RIRFKSNTTSKIRWQQQKQQHLVTAAAFSPDDALFAVACGRS-IQLWHSPSAATNYQLHL 92
R+R + TS + + V + AFSP A G + ++ W + ++
Sbjct 90 RVRAVARCTSSL----EGHTEAVISVAFSPTGKYLASGSGDTTVRFWDLSTETPHH---- 141
Query 93 LTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWR-ARGKWGGQT 137
+ H + ++WSPD L SG ++ + LW GK G+T
Sbjct 142 --TSRGHTHWVLSIAWSPDGKKLASGCKNSQIFLWDPVTGKQIGKT 185
> dre:394014 wdr1, MGC55793, wu:fa66e09, zgc:55793, zgc:77547;
WD repeat domain 1
Length=606
Score = 45.1 bits (105), Expect = 2e-04, Method: Composition-based stats.
Identities = 38/123 (30%), Positives = 52/123 (42%), Gaps = 3/123 (2%)
Query 5 IRSDSQLAISIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSPD 64
I S++ S+D+ Q L G+ N F K + V FSPD
Sbjct 142 IVGHSKIINSVDIK-QTRPYRLVTGSDDNCTTFLEGPPFKFKCTMTDHSRFVNCVRFSPD 200
Query 65 DALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTV 124
+ +A A G Q++ T +L L H I +SWSPDS L+S S D TV
Sbjct 201 GSRYASA-GADGQIFLY-DGKTGEKLSSLGGEKAHDGGIYAVSWSPDSTQLISASGDRTV 258
Query 125 RLW 127
+LW
Sbjct 259 KLW 261
Score = 40.4 bits (93), Expect = 0.004, Method: Composition-based stats.
Identities = 23/77 (29%), Positives = 35/77 (45%), Gaps = 4/77 (5%)
Query 53 QHLVTAAAFSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDS 112
Q VT ++S D A AV + + + + A + + F H + CLSWSPD+
Sbjct 489 QGPVTDMSYSKDGAFLAVTDEKKVVTVFTVADAYKEK----SEFYGHHAKVVCLSWSPDN 544
Query 113 LHLVSGSQDCTVRLWRA 129
H + D V +W A
Sbjct 545 EHFATSGMDMMVYVWTA 561
> hsa:57418 WDR18, MGC2436, R32184_1; WD repeat domain 18
Length=432
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 46/111 (41%), Gaps = 17/111 (15%)
Query 26 LAKGNVLNRIRFKSNTTSKIRWQQQKQQHL---------VTAAAFSPDDALFAVACGRSI 76
L G L + N S W+ Q++ L VT SP+ SI
Sbjct 46 LLNGEYLLAAQLGKNYISA--WELQRKDQLQQKIMCPGPVTCLTASPNGLYVLAGVAESI 103
Query 77 QLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
LW + +LL + H Q ++CL ++ DS H +SG +DC V +W
Sbjct 104 HLWEVSTG------NLLVILSRHYQDVSCLQFTGDSSHFISGGKDCLVLVW 148
Score = 37.7 bits (86), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 28/78 (35%), Positives = 35/78 (44%), Gaps = 4/78 (5%)
Query 96 FTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRARGKWGGQTSAAAAAAAATAATAGTEE 155
F H+ +TCLS S D L+SGS D TVRLW + K +T A A
Sbjct 265 FKGHRNQVTCLSVSTDGSVLLSGSHDETVRLWDVQSKQCIRT----VALKGPVTNAAILL 320
Query 156 AAAAAGDAAFIPSAFLSH 173
A + + F PS L H
Sbjct 321 APVSMLSSDFRPSLPLPH 338
> xla:734559 wdr18, MGC115442; WD repeat domain 18; K14829 pre-rRNA-processing
protein IPI3
Length=428
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/90 (30%), Positives = 40/90 (44%), Gaps = 15/90 (16%)
Query 47 WQQQKQQHL---------VTAAAFSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFT 97
W+ Q++ L V A SP+ SI LW + HLL
Sbjct 65 WELQRRDQLQQKIVCPGPVNCLAASPNGLYLVAGIAESIYLWEVCTG------HLLAILN 118
Query 98 LHQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
H Q +TCL+++ D H+VSG++D V +W
Sbjct 119 NHYQDVTCLTFTDDGSHIVSGAKDSLVLVW 148
Score = 37.0 bits (84), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 40/85 (47%), Gaps = 11/85 (12%)
Query 56 VTAAAFSPDDALFAVACGRS------IQLWHSPSAATNYQLHLLTSFTL--HQQLITCLS 107
+ + AF P A + + CG S + L+ +P Q H L H+ +TCLS
Sbjct 218 IMSVAFDP--AEYHLFCGGSDGVIYQVDLYTTPEQRER-QFHPEQEMVLKGHRNQVTCLS 274
Query 108 WSPDSLHLVSGSQDCTVRLWRARGK 132
S D L+SGS D TV +W + K
Sbjct 275 VSLDGSMLISGSHDETVCVWDIQSK 299
> ath:AT2G47990 SWA1; SWA1 (SLOW WALKER1); nucleotide binding;
K14549 U3 small nucleolar RNA-associated protein 15
Length=530
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 36/132 (27%), Positives = 60/132 (45%), Gaps = 21/132 (15%)
Query 1 STFCIRSDSQLAISIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAA 60
S+ C RSD L + D+ G V ++ + L +R S +++ Q + HLV+
Sbjct 97 SSVCFRSDGALFAACDLSGVVQVFDIKERMALRTLRSHSAPARFVKYPVQDKLHLVSGG- 155
Query 61 FSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSP--DSLHLVSG 118
DD + ++ W A +++ H+ + C SP DS+ LV+G
Sbjct 156 ---DDGV--------VKYWDVAGAT------VISDLLGHKDYVRCGDCSPVNDSM-LVTG 197
Query 119 SQDCTVRLWRAR 130
S D TV++W AR
Sbjct 198 SYDHTVKVWDAR 209
> dre:406291 katnb1, wu:fj32f02, wu:fj65h01, zgc:56071; katanin
p80 (WD repeat containing) subunit B 1
Length=694
Score = 43.9 bits (102), Expect = 4e-04, Method: Composition-based stats.
Identities = 34/115 (29%), Positives = 51/115 (44%), Gaps = 9/115 (7%)
Query 14 SIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSPDDALFAVACG 73
S+D H G LA G+V + I+ ++ + V AFSPD A A
Sbjct 110 SLDFHPMGEY--LASGSVDSNIKLWDVRRKGCVFRYKGHTQAVRCLAFSPDGKWLASASD 167
Query 74 RS-IQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
S ++LW + ++T FT H + + + P+ L SGS D TV+LW
Sbjct 168 DSTVKLWDLIAG------KMITEFTSHTSAVNVVQFHPNEYLLASGSADRTVKLW 216
Score = 34.3 bits (77), Expect = 0.23, Method: Composition-based stats.
Identities = 32/133 (24%), Positives = 57/133 (42%), Gaps = 16/133 (12%)
Query 62 SPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQD 121
S ++ + A + S++LW +A +L + H+ I+ L + P +L SGS D
Sbjct 73 SSEERVVAGSLSGSLRLWDLEAA------KILRTLMGHKASISSLDFHPMGEYLASGSVD 126
Query 122 CTVRLWRARGK-----WGGQTSAAAAAAAATAA---TAGTEEAAAAAGD--AAFIPSAFL 171
++LW R K + G T A A + + ++++ D A + + F
Sbjct 127 SNIKLWDVRRKGCVFRYKGHTQAVRCLAFSPDGKWLASASDDSTVKLWDLIAGKMITEFT 186
Query 172 SHRHELKFVCFSP 184
SH + V F P
Sbjct 187 SHTSAVNVVQFHP 199
> bbo:BBOV_III001050 17.m07120; WD domain, G-beta repeat containing
protein
Length=370
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 45/100 (45%), Gaps = 18/100 (18%)
Query 50 QKQQHLVTAAAFSPDDALFAVACGRSIQLW-HSPSAAT----------------NYQLHL 92
+ ++ V A+F + CGR +W H S++T + +
Sbjct 138 EGHENEVKCASFDCT-GTYVATCGRDKTIWIHQRSSSTPGDTSDIVRSLSGTEGSIDFYC 196
Query 93 LTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRARGK 132
T H Q + C+SWSP +L LVSGS D ++RLW G+
Sbjct 197 AAILTSHSQDVKCVSWSPTALLLVSGSYDNSIRLWGLVGQ 236
Score = 30.0 bits (66), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 89 QLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWR 128
+H L+ H + I + WSP + S DC+V++WR
Sbjct 2 DVHHLSCVKGHSERIWSVCWSPVDDIFATCSSDCSVKIWR 41
> hsa:54475 NLE1, FLJ10458, Nle; notchless homolog 1 (Drosophila);
K14855 ribosome assembly protein 4
Length=193
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 40/81 (49%), Gaps = 7/81 (8%)
Query 53 QHLVTAAAFSPDDALFAVAC-GRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPD 111
Q L+ FSPD + A A +SI+LW + L S H + ++WS D
Sbjct 80 QALINQVLFSPDSRIVASASFDKSIKLWDGRTG------KYLASLRGHVAAVYQIAWSAD 133
Query 112 SLHLVSGSQDCTVRLWRARGK 132
S LVSGS D T+++W + +
Sbjct 134 SRLLVSGSSDSTLKVWDVKAQ 154
Score = 35.0 bits (79), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 46/107 (42%), Gaps = 10/107 (9%)
Query 93 LTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRAR-GKWGGQTSAAAAAAAATAATA 151
LT T HQ LI + +SPDS + S S D +++LW R GK+ AA A +A
Sbjct 73 LTRMTGHQALINQVLFSPDSRIVASASFDKSIKLWDGRTGKYLASLRGHVAAVYQIAWSA 132
Query 152 GTEEAAAAAGDAAF---------IPSAFLSHRHELKFVCFSPCLTRI 189
+ + + D+ + H E+ V +SP R+
Sbjct 133 DSRLLVSGSSDSTLKVWDVKAQKLAMDLPGHADEVYAVDWSPDGQRV 179
> ath:AT2G45540 WD-40 repeat family protein / beige-related
Length=2946
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 41/116 (35%), Positives = 56/116 (48%), Gaps = 13/116 (11%)
Query 75 SIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRARGKWG 134
SI+L S A T L T+F H +TCL+ SPD+ LV+GS+D TV LWR +
Sbjct 2649 SIKLVSSDGAKT-----LETAFG-HCAPVTCLALSPDNNFLVTGSRDSTVLLWRIHKAFT 2702
Query 135 GQTSAA--AAAAAATAATAGTEEAAAAAGDAAFI----PSAFL-SHRHELKFVCFS 183
+TS + + + A ++T+ T A A P L HR EL C S
Sbjct 2703 SRTSVSEPSTGSGAPSSTSNTNLANTLANKGKKCRLEGPIQVLRGHRRELVCCCVS 2758
> mmu:217011 Nle1, AL022765, BC018399, MGC25690, Nle; notchless
homolog 1 (Drosophila); K14855 ribosome assembly protein 4
Length=485
Score = 43.5 bits (101), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 35/140 (25%), Positives = 64/140 (45%), Gaps = 17/140 (12%)
Query 1 STFCIRSDSQLAISIDVHGQGLV--------INLAKGNVLNRIRFKSNTTSKIRWQQQKQ 52
+ F +R+ ++ S++ H + ++ LA G+ +RF +T + +
Sbjct 96 AVFRVRAVTRCTSSLEGHSEAVISVAFSPTGKYLASGSGDTTVRFWDLSTETPHFTCKGH 155
Query 53 QHLVTAAAFSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSW---- 108
+H V + ++SPD A C L PS L + + T H + IT LSW
Sbjct 156 RHWVLSISWSPDGKKLASGCKNGQVLLWDPSTG----LQVGRTLTGHSKWITGLSWEPLH 211
Query 109 -SPDSLHLVSGSQDCTVRLW 127
+P+ ++ S S+D +VR+W
Sbjct 212 MNPECRYVASSSKDGSVRVW 231
Score = 43.1 bits (100), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 33/119 (27%), Positives = 51/119 (42%), Gaps = 17/119 (14%)
Query 25 NLAKGNVLNRIRFKSNTTSKIRW----------QQQKQQHLVTAAAFSPDDALFAVAC-G 73
NL +G R+ S+ + W + Q L+ FSPD + A A
Sbjct 334 NLVRGQGPERLVSGSDDFTLFLWSPAEDKKPLARMTGHQALINQVLFSPDSRIVASASFD 393
Query 74 RSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRARGK 132
+SI+LW + L S H + ++WS DS LVSGS D T+++W + +
Sbjct 394 KSIKLWDGRTG------KYLASLRGHVAAVYQIAWSADSRLLVSGSSDSTLKVWDVKAQ 446
Score = 37.4 bits (85), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 18/34 (52%), Positives = 22/34 (64%), Gaps = 1/34 (2%)
Query 96 FTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRA 129
T H Q +TCL W D L L S SQD T+++WRA
Sbjct 242 LTGHTQSVTCLRWGGDGL-LYSASQDRTIKVWRA 274
Score = 33.5 bits (75), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 46/107 (42%), Gaps = 10/107 (9%)
Query 93 LTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRAR-GKWGGQTSAAAAAAAATAATA 151
L T HQ LI + +SPDS + S S D +++LW R GK+ AA A +A
Sbjct 365 LARMTGHQALINQVLFSPDSRIVASASFDKSIKLWDGRTGKYLASLRGHVAAVYQIAWSA 424
Query 152 GTEEAAAAAGDAAF---------IPSAFLSHRHELKFVCFSPCLTRI 189
+ + + D+ + + H E+ V +SP R+
Sbjct 425 DSRLLVSGSSDSTLKVWDVKAQKLATDLPGHADEVYAVDWSPDGQRV 471
> mmu:22388 Wdr1, Aip1, D5Wsu185e, rede; WD repeat domain 1
Length=606
Score = 43.1 bits (100), Expect = 5e-04, Method: Composition-based stats.
Identities = 34/114 (29%), Positives = 46/114 (40%), Gaps = 3/114 (2%)
Query 14 SIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSPDDALFAVACG 73
S+D+ Q LA G+ N F K ++ V FSPD FA A
Sbjct 151 SVDIK-QTRPYRLATGSDDNCAAFFEGPPFKFKFTIGDHSRFVNCVRFSPDGNRFATASA 209
Query 74 RSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
+ T ++ L H I +SWSPDS HL+S S D T ++W
Sbjct 210 DGQIFIYD--GKTGEKVCALGESKAHDGGIYAISWSPDSTHLLSASGDKTSKIW 261
Score = 42.0 bits (97), Expect = 0.001, Method: Composition-based stats.
Identities = 28/72 (38%), Positives = 35/72 (48%), Gaps = 4/72 (5%)
Query 56 VTAAAFSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHL 115
VT A+S D A AV C S ++ S A Y + F H I CL+WSPD+ H
Sbjct 492 VTDVAYSHDGAFLAV-CDAS-KVVTVFSVADGYSEN--NVFYGHHAKIVCLAWSPDNEHF 547
Query 116 VSGSQDCTVRLW 127
SG D V +W
Sbjct 548 ASGGMDMMVYVW 559
> mmu:58523 Elp2, AU023723, Epl2, StIP1, Statip1; elongation protein
2 homolog (S. cerevisiae); K11374 elongator complex protein
2
Length=831
Score = 42.7 bits (99), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 40/80 (50%), Gaps = 8/80 (10%)
Query 56 VTAAAFSPDDA-LFAVACGRSIQLWHSPSAATNYQLHLLTSF-------TLHQQLITCLS 107
VT FSPDD L AV+ R+ LW A ++ + F ++H ++I
Sbjct 617 VTQMTFSPDDKFLLAVSRDRTWSLWKRQDATSSEFDPFFSLFAFTNKITSVHSRIIWSCD 676
Query 108 WSPDSLHLVSGSQDCTVRLW 127
WSPD+ + +GS+D V +W
Sbjct 677 WSPDNKYFFTGSRDKKVVVW 696
> hsa:6801 STRN, MGC125642, SG2NA; striatin, calmodulin binding
protein
Length=780
Score = 42.4 bits (98), Expect = 8e-04, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 92 LLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRARGK 132
L+ S H + +T L+ P+ L+L+SGS DC++RLW K
Sbjct 698 LIHSMVAHLEAVTSLAVDPNGLYLMSGSHDCSIRLWNLESK 738
> hsa:55250 ELP2, FLJ10879, SHINC-2, STATIP1, StIP; elongation
protein 2 homolog (S. cerevisiae); K11374 elongator complex
protein 2
Length=826
Score = 42.4 bits (98), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 42/80 (52%), Gaps = 8/80 (10%)
Query 56 VTAAAFSPDDA-LFAVACGRSIQLWH-----SPSAATNYQLHLLTS--FTLHQQLITCLS 107
VT AFSP++ L AV+ R+ LW SP + L T+ ++H ++I
Sbjct 617 VTQMAFSPNEKFLLAVSRDRTWSLWKKQDTISPEFEPVFSLFAFTNKITSVHSRIIWSCD 676
Query 108 WSPDSLHLVSGSQDCTVRLW 127
WSPDS + +GS+D V +W
Sbjct 677 WSPDSKYFFTGSRDKKVVVW 696
Score = 32.3 bits (72), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 34/76 (44%), Gaps = 3/76 (3%)
Query 55 LVTAAAFSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSP--DS 112
L +F P+ + +ACG H A N Q + S H+ I + W+
Sbjct 163 LALCLSFLPNTDVPILACGNDDCRIHI-FAQQNDQFQKVLSLCGHEDWIRGVEWAAFGRD 221
Query 113 LHLVSGSQDCTVRLWR 128
L L S SQDC +R+W+
Sbjct 222 LFLASCSQDCLIRIWK 237
> mmu:268980 Strn, AU022939, D130055P19; striatin, calmodulin
binding protein
Length=780
Score = 42.4 bits (98), Expect = 9e-04, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 92 LLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRARGK 132
L+ S H + +T L+ P+ L+L+SGS DC++RLW K
Sbjct 698 LIHSMVAHLEAVTSLAVDPNGLYLMSGSHDCSIRLWNLESK 738
> cel:K08F9.2 tag-216; Temporarily Assigned Gene name family member
(tag-216)
Length=600
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 50/97 (51%), Gaps = 7/97 (7%)
Query 37 FKSNTTSKIRWQQQKQQH--LVTAAAFSPDDALFAVA-CGRSIQLWHSPSAATNYQLHLL 93
+K N ++ + +K +H +TA +FS D AV R + + S +T++ +
Sbjct 463 YKLNGDGRLE-ELKKIEHSAEITAVSFSDDGEYLAVTDLARKVIPY---SVSTDFSVTSP 518
Query 94 TSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRAR 130
S+T H + ++WSPD+ L +GS D +V +W +
Sbjct 519 NSWTFHTSKVLTVAWSPDNQRLATGSIDTSVIIWDMK 555
> dre:791160 strn; zgc:158357
Length=782
Score = 42.4 bits (98), Expect = 0.001, Method: Composition-based stats.
Identities = 18/41 (43%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 92 LLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRARGK 132
L+ S H +T L+ P+ L+L+SGS DC+VRLW K
Sbjct 700 LIHSMVAHLDAVTSLAVDPNGLYLMSGSHDCSVRLWNMESK 740
> ath:AT1G24130 transducin family protein / WD-40 repeat family
protein
Length=415
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 55/110 (50%), Gaps = 18/110 (16%)
Query 37 FKSNTTSKIR-WQQQKQQH-----------LVTAAAFSPD-DALFAVACGRSIQLWHSPS 83
+ + KI+ W ++ ++H V A A S D L++ AC RSI +W
Sbjct 250 YTGSADKKIKVWNKKDKKHSLVATLTKHLSAVNALAISEDGKVLYSGACDRSILVWERLI 309
Query 84 AATNYQLHLLTSFTL--HQQLITCLSWSPDSLHLVSGSQDCTVRLWRARG 131
+ +LH+ L H++ I CL+ + D ++SGS D ++R+WR RG
Sbjct 310 NGDDEELHMSVVGALRGHRKAIMCLAVASDL--VLSGSADKSLRVWR-RG 356
> ath:AT5G64630 FAS2; FAS2 (FASCIATA 2); nucleotide binding /
protein binding; K10751 chromatin assembly factor 1 subunit
B
Length=397
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 36/73 (49%), Gaps = 1/73 (1%)
Query 56 VTAAAFSPDDALFAV-ACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLH 114
V FSP L A A G + +W + TN + S + H++ + L WSPD +
Sbjct 67 VNTIRFSPSGELLASGADGGELFIWKLHPSETNQSWKVHKSLSFHRKDVLDLQWSPDDAY 126
Query 115 LVSGSQDCTVRLW 127
L+SGS D + +W
Sbjct 127 LISGSVDNSCIIW 139
> dre:562780 strn4; striatin, calmodulin binding protein 4
Length=765
Score = 42.0 bits (97), Expect = 0.001, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 92 LLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
++ S H +TCL+ P +L+SGS DC+VRLW
Sbjct 683 VIHSMVAHLDAVTCLATDPKGTYLISGSHDCSVRLW 718
> xla:447445 strn, MGC81438; striatin, calmodulin binding protein
Length=791
Score = 41.6 bits (96), Expect = 0.002, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 92 LLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRARGK 132
L+ S H +T L+ P+ L+L+SGS DC++RLW K
Sbjct 709 LIHSMVAHLDAVTSLAVDPNGLYLMSGSHDCSIRLWNLESK 749
> xla:379542 bop1-a, MGC68939; ribosome biogenesis protein bop1-A;
K14824 ribosome biogenesis protein ERB1
Length=728
Score = 41.6 bits (96), Expect = 0.002, Method: Composition-based stats.
Identities = 16/29 (55%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 99 HQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
H+ L+ C+S SP LVSGS DC+VR W
Sbjct 394 HKDLVRCISVSPSGQWLVSGSDDCSVRFW 422
> xla:380050 bop1, MGC52658, bop1-B; block of proliferation 1;
K14824 ribosome biogenesis protein ERB1
Length=728
Score = 41.6 bits (96), Expect = 0.002, Method: Composition-based stats.
Identities = 16/29 (55%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 99 HQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
H+ L+ C+S SP LVSGS DC+VR W
Sbjct 394 HKDLVRCISVSPSGQWLVSGSDDCSVRFW 422
> xla:779163 wsb2, MGC83316; WD repeat and SOCS box containing
2; K10342 WD repeat and SOCS box-containing protein 2
Length=409
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 41/134 (30%), Positives = 57/134 (42%), Gaps = 28/134 (20%)
Query 16 DVHGQGLVINLA-KGNVLNRIRFKSN------TTSKIR----WQQQKQQHL--------- 55
DVH GL+ NLA +V+ + F +N T S+ + W +K L
Sbjct 141 DVHTGGLLFNLAGHQDVVRDVSFSTNDSLILVTASRDKTLRVWDLKKDGKLLHVLSGHTN 200
Query 56 -VTAAAFSPDDALFAVACGR-SIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSL 113
V A SPD ++ A G S+ LW S L+ HQ + +SPDS
Sbjct 201 WVYCCAISPDCSMLCSAAGENSVLLWSMRSYT------LIRKLEGHQNSVVSCDFSPDSA 254
Query 114 HLVSGSQDCTVRLW 127
LV+ S D TV +W
Sbjct 255 LLVTASYDTTVIMW 268
> ath:AT3G49180 RID3; RID3 (ROOT INITIATION DEFECTIVE 3); nucleotide
binding
Length=438
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 34/72 (47%), Gaps = 17/72 (23%)
Query 56 VTAAAFSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHL 115
+ S D L+ VA G+ ++ WH H + +TCL +S D L
Sbjct 93 LVGGGISGDIYLWEVATGKLLKKWHG-----------------HYRSVTCLVFSGDDSLL 135
Query 116 VSGSQDCTVRLW 127
VSGSQD ++R+W
Sbjct 136 VSGSQDGSIRVW 147
Score = 38.5 bits (88), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 47/108 (43%), Gaps = 15/108 (13%)
Query 23 VINLAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSPDDALFAVACGRSIQLWHSP 82
V +L++G +L I F S ++ A A P +F S +
Sbjct 201 VWSLSRGKLLKNIIFPS---------------VINALALDPGGCVFYAGARDSKIYIGAI 245
Query 83 SAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRAR 130
+A + Y +L S + + ITCL++ D L+SGS+D V +W +
Sbjct 246 NATSEYGTQVLGSVSEKGKAITCLAYCADGNLLISGSEDGVVCVWDPK 293
> dre:393168 smu1, MGC56147, zgc:56147; smu-1 suppressor of mec-8
and unc-52 homolog (C. elegans); K13111 WD40 repeat-containing
protein SMU1
Length=513
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/115 (24%), Positives = 53/115 (46%), Gaps = 10/115 (8%)
Query 21 GLVINLAKGNV----LNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSPD-DALFAVACGRS 75
G+ I+L +G + RF + I++ Q+ V A FSPD L +
Sbjct 181 GMTIDLFRGKAAVKDVEEERFPTQLARHIKFGQKSH---VECARFSPDGQYLVTGSVDGF 237
Query 76 IQLWHSPSAATNYQLHLL--TSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWR 128
I++W+ + L +F + + C+S+S D+ L +G+QD +++W+
Sbjct 238 IEVWNFTTGKIRKDLKYQAQDNFMMMDDAVLCMSFSRDTEMLATGAQDGKIKVWK 292
Score = 32.7 bits (73), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 34/110 (30%), Positives = 50/110 (45%), Gaps = 14/110 (12%)
Query 26 LAKGNVLNRIRFKSNTTSKIR----WQQQKQQHLVTAA----AFSPDDALFAV-ACGRSI 76
L G+V I + TT KIR +Q Q ++ A +FS D + A A I
Sbjct 229 LVTGSVDGFIEVWNFTTGKIRKDLKYQAQDNFMMMDDAVLCMSFSRDTEMLATGAQDGKI 288
Query 77 QLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRL 126
++W S L H + +TCLS+S DS ++S S D T+R+
Sbjct 289 KVWKIQSGQC-----LRRYERAHSKGVTCLSFSKDSTQILSASFDQTIRI 333
Score = 30.4 bits (67), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 16/62 (25%), Positives = 28/62 (45%), Gaps = 6/62 (9%)
Query 93 LTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRARGKWGGQTSAAAAAAAATAATAG 152
L F H + + +PD H +S S D TV++W + T+ + + +AG
Sbjct 342 LKEFRGHSSFVNEATLTPDGHHAISASSDGTVKVWNMK------TTECTSTFKSLGTSAG 395
Query 153 TE 154
T+
Sbjct 396 TD 397
> pfa:PF11_0471 nucleolar preribosomal assembly protein, putative;
K14855 ribosome assembly protein 4
Length=645
Score = 41.2 bits (95), Expect = 0.002, Method: Composition-based stats.
Identities = 17/29 (58%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 99 HQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
H I CL++SP+S HL +GS D TVRLW
Sbjct 124 HTSSILCLAFSPNSSHLATGSGDNTVRLW 152
Score = 36.6 bits (83), Expect = 0.060, Method: Composition-based stats.
Identities = 25/78 (32%), Positives = 39/78 (50%), Gaps = 7/78 (8%)
Query 52 QQHLVTAAAFSPDDALFAVAC-GRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSP 110
Q V A FSP+ + + +SI++W SAA L + H + ++WS
Sbjct 478 HQKPVIHAQFSPNGKMIVSSSFDKSIRVW---SAADG---KFLAVYRGHVGPVYKVAWSI 531
Query 111 DSLHLVSGSQDCTVRLWR 128
D+ VS SQD T++LW+
Sbjct 532 DNNFFVSCSQDSTLKLWK 549
Score = 35.0 bits (79), Expect = 0.17, Method: Composition-based stats.
Identities = 30/126 (23%), Positives = 58/126 (46%), Gaps = 19/126 (15%)
Query 4 CIRSDSQLAISIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSP 63
C+ +S+ + ++ ++IN K N++N I KS K ++ Q + +V+ S
Sbjct 402 CLTLNSERLLKNGIYNLDVIIN--KINIVNYIE-KSKNIMKNFFKNQHHEKIVSG---SD 455
Query 64 DDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCT 123
D LF + C ++ + + T HQ+ + +SP+ +VS S D +
Sbjct 456 DGTLFLIECLQNKEYKN-------------TRLLGHQKPVIHAQFSPNGKMIVSSSFDKS 502
Query 124 VRLWRA 129
+R+W A
Sbjct 503 IRVWSA 508
Score = 29.6 bits (65), Expect = 7.1, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 20/35 (57%), Gaps = 3/35 (8%)
Query 96 FTLHQQLITCLSWS---PDSLHLVSGSQDCTVRLW 127
T H ITC+ WS ++ L S S+D T+++W
Sbjct 348 LTGHSDTITCILWSGRDTENSTLYSSSRDTTIKIW 382
> dre:445819 wdr18, wu:fa16f06, wu:fb79d07, zgc:92370; WD repeat
domain 18; K14829 pre-rRNA-processing protein IPI3
Length=431
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/60 (43%), Positives = 32/60 (53%), Gaps = 8/60 (13%)
Query 96 FTLHQQLITCLSWSPDSLHLVSGSQDCTVRLWRARGK---W-----GGQTSAAAAAAAAT 147
F H+ L+TCLS S D L+SGS D TVR+W + K W G T+AA A A
Sbjct 265 FKGHRNLVTCLSVSMDGTVLLSGSNDETVRMWDVQSKQCIWTINHRGPVTNAAIIPAPAN 324
Score = 38.5 bits (88), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 25/90 (27%), Positives = 40/90 (44%), Gaps = 11/90 (12%)
Query 44 KIRWQQQKQQHLV-----TAAAFSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTL 98
+I+ + Q QQ +V T SPD +I LW + +LL +
Sbjct 66 EIQRKDQLQQKIVCPGIVTCLCASPDGLYVLAGIAEAIYLWEVSTG------NLLAILSR 119
Query 99 HQQLITCLSWSPDSLHLVSGSQDCTVRLWR 128
H Q ++C+ ++ DS H VSG +D +W
Sbjct 120 HFQDLSCIKFTDDSSHFVSGGKDNLAFIWN 149
> tgo:TGME49_015740 hypothetical protein ; K14855 ribosome assembly
protein 4
Length=527
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 38/80 (47%), Gaps = 7/80 (8%)
Query 52 QQHLVTAAAFSPDDALFAVAC-GRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSP 110
Q LV AFSPD A A +SI+LW L++ H + L+WS
Sbjct 413 HQKLVNHVAFSPDGRYIASASFDKSIRLWDGRRGV------YLSTLRGHVGPVYQLAWSS 466
Query 111 DSLHLVSGSQDCTVRLWRAR 130
DS L+S S D T+++W A
Sbjct 467 DSRLLLSASGDSTLKVWHAE 486
Score = 39.7 bits (91), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 18/34 (52%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 94 TSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
TS H + I C+++SPD L SGS D TVRLW
Sbjct 137 TSLEGHTEAILCVAFSPDGSQLASGSGDMTVRLW 170
> hsa:317 APAF1, APAF-1, CED4, DKFZp781B1145; apoptotic peptidase
activating factor 1; K02084 apoptotic protease-activating
factor
Length=1194
Score = 40.8 bits (94), Expect = 0.003, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 48/94 (51%), Gaps = 10/94 (10%)
Query 37 FKSNTTSKIRWQQQKQQHLVTAAAFSPDDALFAV-ACGRSIQLWHSPSAATNYQLHLLTS 95
FK+ T K+ + + + V AFS DD A + + +++W+S + L+ +
Sbjct 631 FKAETGEKL-LEIKAHEDEVLCCAFSTDDRFIATCSVDKKVKIWNSMTG------ELVHT 683
Query 96 FTLHQQLITCLSWSPDSLHLV--SGSQDCTVRLW 127
+ H + + C ++ S HL+ +GS DC ++LW
Sbjct 684 YDEHSEQVNCCHFTNSSHHLLLATGSSDCFLKLW 717
Score = 34.7 bits (78), Expect = 0.19, Method: Composition-based stats.
Identities = 21/82 (25%), Positives = 37/82 (45%), Gaps = 6/82 (7%)
Query 49 QQKQQHLVTAAAFSPDDALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSW 108
Q+ + +V ++S D A VA I LW++ S + + H + + +
Sbjct 783 QEDMEVIVKCCSWSADGARIMVAAKNKIFLWNTDSRSK------VADCRGHLSWVHGVMF 836
Query 109 SPDSLHLVSGSQDCTVRLWRAR 130
SPD ++ S D T+RLW +
Sbjct 837 SPDGSSFLTSSDDQTIRLWETK 858
> ath:AT5G08560 transducin family protein / WD-40 repeat family
protein
Length=589
Score = 40.4 bits (93), Expect = 0.003, Method: Composition-based stats.
Identities = 23/85 (27%), Positives = 37/85 (43%), Gaps = 9/85 (10%)
Query 46 RWQQQKQQHLVTAAAFSPDDALFAVACGRSIQ--LWHSPSAATNYQLHLLTSFTLHQQLI 103
R++ K+ + + F F + Q +WH + L+ H +
Sbjct 481 RYKGHKRSRFIIRSCFGGYKQAFIASGSEDSQVYIWHRSTG------KLIVELPGHAGAV 534
Query 104 TCLSWSPDSLH-LVSGSQDCTVRLW 127
C+SWSP +LH L S S D T+R+W
Sbjct 535 NCVSWSPTNLHMLASASDDGTIRIW 559
> cel:F55F8.3 hypothetical protein; K14558 periodic tryptophan
protein 2
Length=910
Score = 40.4 bits (93), Expect = 0.004, Method: Composition-based stats.
Identities = 29/114 (25%), Positives = 54/114 (47%), Gaps = 8/114 (7%)
Query 13 ISIDVHGQGLVINLAKGNVLNRIRFKSNTTSKIRWQQQKQQHLVTAAAFSPDDALFAV-A 71
++ ++ G L I KG+ + ++ + S + +QQ +T A +SPD +L A A
Sbjct 321 VAWNLTGDWLAIGCGKGSTAQLVVWEWQSESYVM-KQQAHSLRITTAEYSPDGSLMATGA 379
Query 72 CGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVR 125
+++W+S S+ +F H +T + W+ ++S S D TVR
Sbjct 380 EDGKVKIWNSRSSFCT------VTFDEHTSGVTAVKWTQSGRAILSASLDGTVR 427
Score = 34.3 bits (77), Expect = 0.28, Method: Composition-based stats.
Identities = 30/124 (24%), Positives = 47/124 (37%), Gaps = 15/124 (12%)
Query 68 FAVACGR--SIQL----WHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQD 121
A+ CG+ + QL W S S Q H L IT +SPD + +G++D
Sbjct 330 LAIGCGKGSTAQLVVWEWQSESYVMKQQAHSLR--------ITTAEYSPDGSLMATGAED 381
Query 122 CTVRLWRARGKWGGQT-SAAAAAAAATAATAGTEEAAAAAGDAAFIPSAFLSHRHELKFV 180
V++W +R + T + A T +A+ D +R+ V
Sbjct 382 GKVKIWNSRSSFCTVTFDEHTSGVTAVKWTQSGRAILSASLDGTVRAHDLKRYRNFRTLV 441
Query 181 CFSP 184
C P
Sbjct 442 CPEP 445
> tpv:TP04_0285 hypothetical protein; K14855 ribosome assembly
protein 4
Length=470
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 38/94 (40%), Gaps = 9/94 (9%)
Query 56 VTAAAFSPDDALFAVACGRS-IQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLH 114
V FSPD A G + +++W + + +FT H + +SWSPD
Sbjct 103 VLCLEFSPDGVYLASGSGDTTVRIWDLATQTP------IKTFTGHTNWVMSISWSPDGYT 156
Query 115 LVSGSQDCTVRLWRARGKWGGQTSAAAAAAAATA 148
L SG D V +W + G T A TA
Sbjct 157 LSSGGMDNKVIIWNPKT--GSGTDLKGHTKAVTA 188
Score = 39.3 bits (90), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 23/34 (67%), Gaps = 0/34 (0%)
Query 94 TSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
+S H + + CL +SPD ++L SGS D TVR+W
Sbjct 94 SSLEGHTESVLCLEFSPDGVYLASGSGDTTVRIW 127
Score = 34.7 bits (78), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 28/46 (60%), Gaps = 3/46 (6%)
Query 82 PSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
P++ +N LH LT HQQLI +S+S + + S S D ++R+W
Sbjct 346 PNSDSNKPLHRLTG---HQQLINHVSFSSNGRYFASASFDKSIRIW 388
Score = 32.3 bits (72), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 37/85 (43%), Gaps = 19/85 (22%)
Query 56 VTAAAFSPDD-ALFAVACGRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLH 114
V + ++SPD L + + +W+ + + T H + +T LSW P LH
Sbjct 145 VMSISWSPDGYTLSSGGMDNKVIIWNPKTGSG-------TDLKGHTKAVTALSWQP--LH 195
Query 115 ---------LVSGSQDCTVRLWRAR 130
L SGS D TVR+W +
Sbjct 196 NLDANEYPLLASGSMDYTVRIWNVK 220
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 37/80 (46%), Gaps = 7/80 (8%)
Query 52 QQHLVTAAAFSPDDALFAVAC-GRSIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSP 110
Q L+ +FS + FA A +SI++W + L + H + ++WS
Sbjct 360 HQQLINHVSFSSNGRYFASASFDKSIRIWCGITGK------YLRTLRGHIGRVYRVAWSC 413
Query 111 DSLHLVSGSQDCTVRLWRAR 130
+LVS S D T++LW A
Sbjct 414 RGNYLVSASSDSTLKLWDAE 433
> mmu:70235 Poc1a, 2510040D07Rik, Wdr51a; POC1 centriolar protein
homolog A (Chlamydomonas)
Length=405
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 31/53 (58%), Gaps = 6/53 (11%)
Query 75 SIQLWHSPSAATNYQLHLLTSFTLHQQLITCLSWSPDSLHLVSGSQDCTVRLW 127
++ +WH + Y+ FT H+ +TC+++SP L SGS+D TVR+W
Sbjct 42 TLMIWHMKPQSRAYR------FTGHKDAVTCVNFSPSGHLLASGSRDKTVRIW 88
Lambda K H
0.322 0.128 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5364689396
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40