bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3442_orf2
Length=226
Score E
Sequences producing significant alignments: (Bits) Value
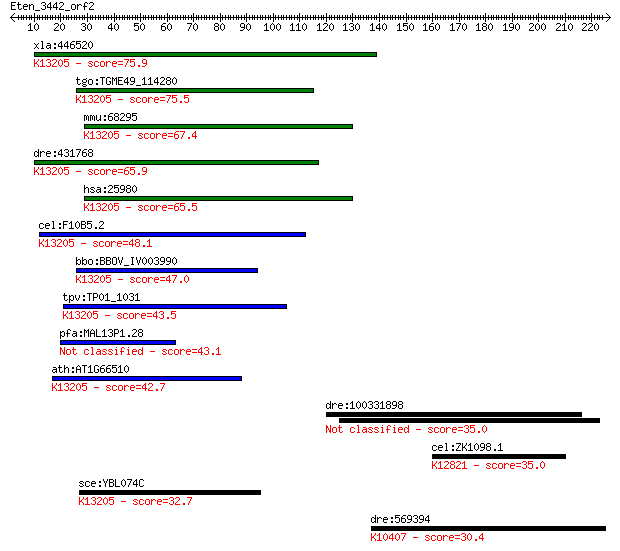
xla:446520 c20orf4, MGC80226; chromosome 20 open reading frame... 75.9 1e-13
tgo:TGME49_114280 hypothetical protein ; K13205 A1 cistron-spl... 75.5 1e-13
mmu:68295 0610011L14Rik; RIKEN cDNA 0610011L14 gene; K13205 A1... 67.4 4e-11
dre:431768 im:7153138; zgc:92018; K13205 A1 cistron-splicing f... 65.9 1e-10
hsa:25980 C20orf4, DKFZp564N1363, bA234K24.2; chromosome 20 op... 65.5 1e-10
cel:F10B5.2 hypothetical protein; K13205 A1 cistron-splicing f... 48.1 3e-05
bbo:BBOV_IV003990 23.m06321; hypothetical protein; K13205 A1 c... 47.0 5e-05
tpv:TP01_1031 hypothetical protein; K13205 A1 cistron-splicing... 43.5 7e-04
pfa:MAL13P1.28 conserved Plasmodium protein, unknown function 43.1 7e-04
ath:AT1G66510 AAR2 protein family; K13205 A1 cistron-splicing ... 42.7 0.001
dre:100331898 eEF1A2 binding protein-like 35.0 0.21
cel:ZK1098.1 hypothetical protein; K12821 pre-mRNA-processing ... 35.0 0.22
sce:YBL074C AAR2; Aar2p; K13205 A1 cistron-splicing factor AAR2 32.7 0.99
dre:569394 klc3, MGC153164, wu:fk22d07, zgc:153164; kinesin li... 30.4 4.7
> xla:446520 c20orf4, MGC80226; chromosome 20 open reading frame
4; K13205 A1 cistron-splicing factor AAR2
Length=388
Score = 75.9 bits (185), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 48/130 (36%), Positives = 69/130 (53%), Gaps = 5/130 (3%)
Query 10 DRSDALLLLIREHQEGFAA-VLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALYL 68
D S AL L++ H G +LAELQ +F+ VLG+ Y + WK LL L+C +E
Sbjct 225 DLSYALEQLLKTHYTGQPLQLLAELQFSFVCFVLGNVYEAFEQWKSLLNLLCRAETFSLQ 284
Query 69 YPELFSKFIDTFYAQLEQAPDELTELEPLQQNNLLTSCCAALLEFCCSVDRQAARAALKQ 128
+PEL+ K I Y QL Q P + ++ + QNN LTS L F CS + A +L++
Sbjct 285 HPELYIKVISVLYHQLAQVPTDFF-VDIVSQNNFLTSTLQVLFSFLCSPN---ANPSLRK 340
Query 129 QQVEAAAAAT 138
+ + A T
Sbjct 341 KAIRFRAHLT 350
> tgo:TGME49_114280 hypothetical protein ; K13205 A1 cistron-splicing
factor AAR2
Length=653
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 34/89 (38%), Positives = 55/89 (61%), Gaps = 1/89 (1%)
Query 26 FAAVLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALYLYPELFSKFIDTFYAQLE 85
+ +L E+Q+A++A +LGH + WK LL L+C+ ERA+ PE++ F+ + QLE
Sbjct 459 WENLLGEIQVAYLAFLLGHRFDGFEQWKQLLLLLCHCERAIRFLPEMYVSFLRLLHCQLE 518
Query 86 QAPDELTELEPLQQNNLLTSCCAALLEFC 114
Q PD+L + L Q + L S +AL++ C
Sbjct 519 QCPDDLLS-DSLLQKSFLFSSASALVDTC 546
> mmu:68295 0610011L14Rik; RIKEN cDNA 0610011L14 gene; K13205
A1 cistron-splicing factor AAR2
Length=384
Score = 67.4 bits (163), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 37/101 (36%), Positives = 54/101 (53%), Gaps = 4/101 (3%)
Query 29 VLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALYLYPELFSKFIDTFYAQLEQAP 88
VL ELQ AF+ +LG+ Y + HWK LL L+C SE A+ Y L+ I Y QL + P
Sbjct 251 VLGELQFAFVCFLLGNVYEAFEHWKRLLNLLCRSESAMGKYHALYISLISILYHQLGEIP 310
Query 89 DELTELEPLQQNNLLTSCCAALLEFCCSVDRQAARAALKQQ 129
+ ++ + Q+N LTS CS+ A A L+++
Sbjct 311 ADFF-VDIVSQDNFLTSTLQVFFSSACSI---AVEATLRKK 347
> dre:431768 im:7153138; zgc:92018; K13205 A1 cistron-splicing
factor AAR2
Length=384
Score = 65.9 bits (159), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 41/108 (37%), Positives = 58/108 (53%), Gaps = 2/108 (1%)
Query 10 DRSDAL-LLLIREHQEGFAAVLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALYL 68
DRS L +L R ++E VL ELQ AF+ ++G+ Y + HWK LL L+C SE A+
Sbjct 231 DRSYTLNAVLERHYKEQPLNVLGELQFAFVCFLVGNVYEAFEHWKSLLALLCRSEEAMKD 290
Query 69 YPELFSKFIDTFYAQLEQAPDELTELEPLQQNNLLTSCCAALLEFCCS 116
EL+ I Y QL + P + L+ + Q+N LTS +F S
Sbjct 291 RKELYLGLIAVLYHQLGEIPPDFF-LDIVSQSNFLTSTLQDFFQFASS 337
> hsa:25980 C20orf4, DKFZp564N1363, bA234K24.2; chromosome 20
open reading frame 4; K13205 A1 cistron-splicing factor AAR2
Length=384
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 54/101 (53%), Gaps = 4/101 (3%)
Query 29 VLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALYLYPELFSKFIDTFYAQLEQAP 88
VL ELQ AF+ +LG+ Y + HWK LL L+C SE A+ + L+ I Y QL + P
Sbjct 251 VLGELQFAFVCFLLGNVYEAFEHWKRLLNLLCRSEAAMMKHHTLYINLISILYHQLGEIP 310
Query 89 DELTELEPLQQNNLLTSCCAALLEFCCSVDRQAARAALKQQ 129
+ ++ + Q+N LTS CS+ A A L+++
Sbjct 311 ADFF-VDIVSQDNFLTSTLQVFFSSACSI---AVDATLRKK 347
> cel:F10B5.2 hypothetical protein; K13205 A1 cistron-splicing
factor AAR2
Length=357
Score = 48.1 bits (113), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 51/100 (51%), Gaps = 1/100 (1%)
Query 12 SDALLLLIREHQEGFAAVLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALYLYPE 71
SD L L+R + +LAE+Q+AF+ + G + WK ++ L+ +L E
Sbjct 224 SDRLYRLLRALGGDWKQLLAEMQIAFVCFLQGQVFEGFEQWKRIIHLMSCCPNSLGSEKE 283
Query 72 LFSKFIDTFYAQLEQAPDELTELEPLQQNNLLTSCCAALL 111
LF FI + QL++ P + ++ + ++N LT+ + L
Sbjct 284 LFMSFIRVLFFQLKECPTDFF-VDIVSRDNFLTTTLSMLF 322
> bbo:BBOV_IV003990 23.m06321; hypothetical protein; K13205 A1
cistron-splicing factor AAR2
Length=523
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 35/68 (51%), Gaps = 0/68 (0%)
Query 26 FAAVLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALYLYPELFSKFIDTFYAQLE 85
+ L E Q AF +L +HY S HWK + +++CN+E L ++ K ++ QLE
Sbjct 396 YGLFLGEYQYAFCVFLLNYHYASFEHWKSMFRILCNAETFLIENVDVGEKVLNIIKHQLE 455
Query 86 QAPDELTE 93
+L +
Sbjct 456 SFESDLYD 463
> tpv:TP01_1031 hypothetical protein; K13205 A1 cistron-splicing
factor AAR2
Length=665
Score = 43.5 bits (101), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 45/84 (53%), Gaps = 3/84 (3%)
Query 21 EHQEGFAAVLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALYLYPELFSKFIDTF 80
E + + VL ELQ +F+ +L ++ SL +K LL+ CN+E L EL K + T
Sbjct 535 EFKNKYFYVLGELQYSFLVFILCFNFESLEQYKRLLRAFCNAESLLVSDQELTQKLLKTL 594
Query 81 YAQLEQAPDELTELEPLQQNNLLT 104
Q+E DE E + Q++N LT
Sbjct 595 RFQIE-TWDE--EHDVYQKDNFLT 615
> pfa:MAL13P1.28 conserved Plasmodium protein, unknown function
Length=609
Score = 43.1 bits (100), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 26/50 (52%), Gaps = 7/50 (14%)
Query 20 REHQEGFAA-------VLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNS 62
E EGF +L E Q AFI LG++Y S + W+ L +L+ NS
Sbjct 461 NEQIEGFVKYSDKLFYILGEFQFAFILFHLGYNYQSFIQWRKLFELMTNS 510
> ath:AT1G66510 AAR2 protein family; K13205 A1 cistron-splicing
factor AAR2
Length=399
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 40/72 (55%), Gaps = 1/72 (1%)
Query 17 LLIREHQEGFAAVLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALY-LYPELFSK 75
+L +E+++ +L ELQ +F+A ++G S + WK ++ L+ A + +LF+K
Sbjct 225 VLSKEYKDSEDLLLGELQFSFVAFLMGQSLESFMQWKSIVSLLLGCTSAPFQTRSQLFTK 284
Query 76 FIDTFYAQLEQA 87
FI Y QL+
Sbjct 285 FIKVIYHQLKYG 296
> dre:100331898 eEF1A2 binding protein-like
Length=1300
Score = 35.0 bits (79), Expect = 0.21, Method: Composition-based stats.
Identities = 36/106 (33%), Positives = 50/106 (47%), Gaps = 15/106 (14%)
Query 120 QAARAALKQQ------QVEAAAAATPAAAAAGKAAPTRRREVHDRHPDAMQTEQTQQQPP 173
+AAR A K+Q Q E AA A A KA+ R+ D+ A + ++ Q +
Sbjct 444 EAARKAKKEQDDARRAQAERDEAARRAKADQDKAS---RKAKADQDEAARKAQEEQDE-- 498
Query 174 AAAEATAPAAEAAAAAQA----AAAEAEIDRAVAARLAALEPQYKE 215
AA + A EAA AQA AA A DR A + AA+ + +E
Sbjct 499 AAKRSQAERDEAARRAQAERDEAARRAHADREAAEKRAAMMAKKEE 544
Score = 32.0 bits (71), Expect = 1.7, Method: Composition-based stats.
Identities = 37/113 (32%), Positives = 51/113 (45%), Gaps = 15/113 (13%)
Query 125 ALKQQQVEAAAAATPAAAAAGKAAPTRRREVHD-RHPDAMQTEQTQQ----QPPAAAEAT 179
A ++ Q E AA A A +AA ++E D R A + E ++ Q A+ +A
Sbjct 423 AARRAQAEKDEAARRAQAERDEAARKAKKEQDDARRAQAERDEAARRAKADQDKASRKAK 482
Query 180 APAAEAAAAAQ------AAAAEAEID----RAVAARLAALEPQYKEVEAAAKR 222
A EAA AQ A ++AE D RA A R A + + EAA KR
Sbjct 483 ADQDEAARKAQEEQDEAAKRSQAERDEAARRAQAERDEAARRAHADREAAEKR 535
> cel:ZK1098.1 hypothetical protein; K12821 pre-mRNA-processing
factor 40
Length=724
Score = 35.0 bits (79), Expect = 0.22, Method: Composition-based stats.
Identities = 19/50 (38%), Positives = 30/50 (60%), Gaps = 1/50 (2%)
Query 160 PDAMQTEQTQQQPPAAAEATAPAAEAAAAAQAAAAEAEIDRAVAARLAAL 209
PD + + +Q+P A A AT AAA Q AE+++D+A+ A LA++
Sbjct 154 PDGEEITKGEQKPAAKA-ATVDTVALAAAVQQKKAESDLDKAMKATLASM 202
> sce:YBL074C AAR2; Aar2p; K13205 A1 cistron-splicing factor AAR2
Length=355
Score = 32.7 bits (73), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 16/69 (23%), Positives = 36/69 (52%), Gaps = 5/69 (7%)
Query 27 AAVLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALYLYPE-LFSKFIDTFYAQLE 85
+ ELQ AF+ + +Y S + W +++L+C+S P+ + K + Y Q++
Sbjct 218 SNYFGELQFAFLNAMFFGNYGSSLQWHAMIELICSSATV----PKHMLDKLDEILYYQIK 273
Query 86 QAPDELTEL 94
P++ +++
Sbjct 274 TLPEQYSDI 282
> dre:569394 klc3, MGC153164, wu:fk22d07, zgc:153164; kinesin
light chain 3; K10407 kinesin light chain
Length=512
Score = 30.4 bits (67), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 22/88 (25%), Positives = 42/88 (47%), Gaps = 1/88 (1%)
Query 137 ATPAAAAAGKAAPTRRREVHDRHPDAMQTEQTQQQPPAAAEATAPAAEAAAAAQAAAAEA 196
+T A +A R + D + +Q+ QT + E T+ E+ + EA
Sbjct 10 STQQVIAGLEALRGENRTLLDSLQETLQS-QTSSESSLEQEKTSIILESLERIELGLGEA 68
Query 197 EIDRAVAARLAALEPQYKEVEAAAKRLC 224
++ A++A L +LE + +++ A +RLC
Sbjct 69 QVMMALSAHLGSLEAEKQKLRAQVRRLC 96
Lambda K H
0.318 0.127 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7504534908
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40