bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3493_orf2
Length=241
Score E
Sequences producing significant alignments: (Bits) Value
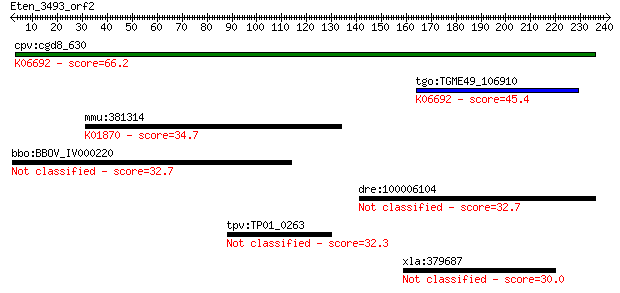
cpv:cgd8_630 26S proteasome regulatory complex, subunit PSMD5 ... 66.2 9e-11
tgo:TGME49_106910 hypothetical protein ; K06692 26S proteasome... 45.4 2e-04
mmu:381314 Iars2, 2010002H18Rik, C79125, MGC63429; isoleucine-... 34.7 0.32
bbo:BBOV_IV000220 21.m02868; hypothetical protein 32.7 1.1
dre:100006104 carcinoembryonic antigen-related cell adhesion m... 32.7 1.1
tpv:TP01_0263 hypothetical protein 32.3 1.7
xla:379687 tfcp2l1-b, crtr1, lbp-9, lbp9, tfcp2l1b; transcript... 30.0 7.0
> cpv:cgd8_630 26S proteasome regulatory complex, subunit PSMD5
; K06692 26S proteasome non-ATPase regulatory subunit 5
Length=505
Score = 66.2 bits (160), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 46/246 (18%), Positives = 106/246 (43%), Gaps = 22/246 (8%)
Query 3 TLLRLLCRMAVERPSECSRILNAEGGVLKSAIETCLTSSSKRHKEE-------GIVCWSI 55
+++ LL +M P +IL G + + S + E I CW +
Sbjct 261 SIVYLLSKMVNNNPELSQKILTMSGNSFVQTFKEFMGSYNSISCSEDYLKSVCAINCWGL 320
Query 56 LFSKTECSATVLEQLPSSADLVAEAVMDVE-DVSVAALRAWSCVLQDVKDPAVLPQQ--- 111
S ++ S L+ + V + + ++S+++L +W + +L Q
Sbjct 321 FASNLISFQNLMNVWEDSFHLMLKLVSNSKSEISLSSLNSWVTFFESKDINQILNQTKTT 380
Query 112 --LKRTVDSNLASKLLDCVLQRPFPEARQQCYELLKVCAMCVVCVSALSVDAEPMQMFFA 169
LK+++++ L ++ ++ +PFPE R Y K+ + + S ++ + +
Sbjct 381 FGLKQSIETQLLPIIISDLISKPFPENRTLIY---KILTLMIPYFSNITSN------LIS 431
Query 170 SEGFRQLMVDPSSDTEYEVRLAKHAFASAACAAQRDWLRNALDPSFCEAFFKYAEGGPWT 229
+ R ++++P+SD+ ++ K+ ++ + N DP+F + +Y + GP+
Sbjct 432 NSKIRDILLNPNSDSNKDILYIKYDLVKQMVKYDQNTINNIPDPNFSSSLQEYVKVGPFY 491
Query 230 ASAAAR 235
S++A
Sbjct 492 KSSSAE 497
> tgo:TGME49_106910 hypothetical protein ; K06692 26S proteasome
non-ATPase regulatory subunit 5
Length=476
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 32/65 (49%), Gaps = 0/65 (0%)
Query 164 MQMFFASEGFRQLMVDPSSDTEYEVRLAKHAFASAACAAQRDWLRNALDPSFCEAFFKYA 223
+Q SE R+ +VDP SD + + AK+ F A DWL ++ S+ +A
Sbjct 396 VQTIMGSEEIRRSLVDPFSDDSSDAKYAKNTFIKKLAADHLDWLGTVIEESYLTKLKTFA 455
Query 224 EGGPW 228
+ GP+
Sbjct 456 DAGPF 460
> mmu:381314 Iars2, 2010002H18Rik, C79125, MGC63429; isoleucine-tRNA
synthetase 2, mitochondrial (EC:6.1.1.5); K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1012
Score = 34.7 bits (78), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 45/111 (40%), Gaps = 8/111 (7%)
Query 31 KSAIETCLTSSSKRH-----KEEGIVCWSILFSKTECSATVLEQLPSSADLVAEAVMDVE 85
K+ E + S + H ++ G W L ++ A VL Q L D+
Sbjct 538 KTKDEYLINSQTTEHIIKLVEQHGSDVWWTLPAEQLLPAEVLAQAGGPGALEYAPGQDIL 597
Query 86 DVSVAALRAWSCVLQDVKDPAVLPQQLKRTVDSNLASKLLDCVLQR---PF 133
D+ + +WSCVLQD + A L + K + S LL V R PF
Sbjct 598 DIWFDSGTSWSCVLQDTQQRADLYLEGKDQLGGWFQSSLLTSVATRSKAPF 648
> bbo:BBOV_IV000220 21.m02868; hypothetical protein
Length=4381
Score = 32.7 bits (73), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 50/115 (43%), Gaps = 15/115 (13%)
Query 2 QTLLRLLCRMAVERPSECSRILNAEGGVLKSAIETCLTSSSKRHKEEGIVCWSILFSKTE 61
+TL RL R A+ P + R E L S+ K++ +V WS + T+
Sbjct 2398 RTLNRLDERGAIMWPEDYKR---------HHKYEYVLRISATIPKDQLLVPWSFVHYSTD 2448
Query 62 CSATVLEQLPSSADLVAEAVMDVEDVSVAALRAWSCVL---QDVKDPAVLPQQLK 113
+ V+E +P V +AV+D + +S V+ + V+ P V P Q K
Sbjct 2449 YTKDVMEMVPCK---VGDAVVDDKFISTMGAEVLGEVIEHREHVEPPKVAPDQKK 2500
> dre:100006104 carcinoembryonic antigen-related cell adhesion
molecule 1-like
Length=389
Score = 32.7 bits (73), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 38/103 (36%), Gaps = 12/103 (11%)
Query 141 YELLKVCAMCVVCVSALSVDAEP------MQMFFASE--GFRQLMVDPSSDTEYEVRLAK 192
Y LK+CA+ ++ P M++F+ + GF L VDP EY RL
Sbjct 29 YSTLKICALQNSTITISCTFTYPTTVHQIMKVFWTKDVNGFPDLSVDP----EYSQRLQY 84
Query 193 HAFASAACAAQRDWLRNALDPSFCEAFFKYAEGGPWTASAAAR 235
C + + + + F GG WT + R
Sbjct 85 LGDKQQICTVRLSHVTKKDEHQYYFRFITDQNGGKWTGTPGVR 127
> tpv:TP01_0263 hypothetical protein
Length=2659
Score = 32.3 bits (72), Expect = 1.7, Method: Composition-based stats.
Identities = 17/44 (38%), Positives = 25/44 (56%), Gaps = 2/44 (4%)
Query 88 SVAALRAWSCVLQDVKDPA--VLPQQLKRTVDSNLASKLLDCVL 129
SV +R WSC ++ + + V+ QLKRTVDS + + VL
Sbjct 1921 SVQNVRRWSCAIKKINKRSGDVIITQLKRTVDSPATMRFKESVL 1964
> xla:379687 tfcp2l1-b, crtr1, lbp-9, lbp9, tfcp2l1b; transcription
factor CP2-like 1
Length=493
Score = 30.0 bits (66), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 24/61 (39%), Gaps = 7/61 (11%)
Query 159 VDAEPMQMFFASEGFRQLMVDPSSDTEYEVRLAKHAFASAACAAQRDWLRNALDPSFCEA 218
VD P++ AS GF DP KH SA+ + WL+ P +C
Sbjct 295 VDPAPLRTDKASTGFNSEYWDPEDKI-------KHLLPSASIQDVQQWLQKNRFPQYCRL 347
Query 219 F 219
F
Sbjct 348 F 348
Lambda K H
0.321 0.129 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8466654768
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40